Combined Multi-Omics Analysis Reveals the Potential Role of ACADS in Yak Intramuscular Fat Deposition
- PMID: 39201818
- PMCID: PMC11354380
- DOI: 10.3390/ijms25169131
Combined Multi-Omics Analysis Reveals the Potential Role of ACADS in Yak Intramuscular Fat Deposition
Abstract
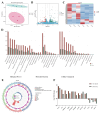
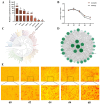
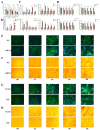
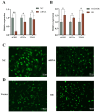
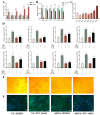
The Yak (Bos grunniens) is a special breed of livestock predominantly distributed in the Qinghai-Tibet Plateau of China. Intramuscular fat (IMF) content in beef cattle is a vital indicator of meat quality. In this study, RNA-Seq and Protein-Seq were respectively employed to sequence the transcriptome and proteome of the longissimus dorsi (LD) tissue from 4-year-old yaks with significant differences in IMF content under the same fattening conditions. Five overlapping genes (MYL3, ACADS, L2HGDH, IGFN1, and ENSBGRG00000000-926) were screened using combined analysis. Functional verification tests demonstrated that the key gene ACADS inhibited yak intramuscular preadipocyte (YIMA) differentiation and proliferation, promoted mitochondrial biogenesis gene expression, and increased the mitochondrial membrane potential (MMP). Furthermore, co-transfection experiments further demonstrated that interfering with ACADS reversed the effect of PPARα agonists in promoting lipid differentiation. In conclusion, ACADS potentially inhibits lipid deposition in YIAMs by regulating the PPARα signalling pathway. These findings offer insights into the molecular mechanisms underlying yak meat quality.
Keywords: ACADS; IMF; PPARα; proteome; transcriptome; yak.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Wang S., Shi J.H., Wang Z.S., Hu R., Wang J.M., Xue B., Peng Q.H. Solation and Identification of preadipocytes from different parts of yak and expression of key genes for differentiation. J. Anim. Sci. Vet. Med. 2022;53:755–765.
-
- Mu Y.P., Li S., Li X.M. Report on Feeding Effect of Jiajiawang Lactic Acid Complex Acidifier. J. Anim. Sci. Vet. Med. 2011;30:7–8.
-
- Hu J.L., Zhuang L., Wu S. Research Progress on Candidate Genes Influencing Meat Quality Traits in Yaks. Chin. J. Anim. Sci. 2021;57:10–15.
-
- Wang Y.L. Master’s Thesis. Yanbian University; Yanbian, China: 2009. Effects of Dietary Energy Level and Source on the Expression of Genes Related to Fat Metabolism in Skeletal Muscle of Finishing Pigs.
-
- Mota de Sá P., Richard A.J., Hang H., Stephens J.M. Transcriptional Regulation of Adipogenesis. Compr. Physiol. 2017;7:635–674. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

