Effects of Curcumin and Estrogen Receptor Alpha in Luminal Breast Cancer Cells
- PMID: 39202273
- PMCID: PMC11353822
- DOI: 10.3390/diagnostics14161785
Effects of Curcumin and Estrogen Receptor Alpha in Luminal Breast Cancer Cells
Abstract
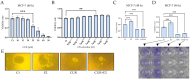
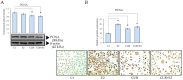
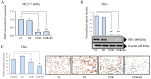
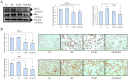
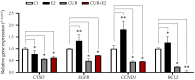
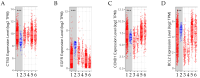
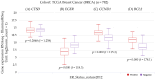
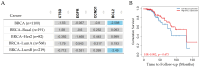
This work examined the potential benefit of curcumin in breast cancer patients as a supplementary drug in ER-positive cancers. The results indicated that in the MCF-7 human breast cancer cell line, E2 and curcumin decreased cell proliferation and the colony-forming capacity and down-regulated protein expression as well as important molecules associated with cell proliferation, such as PCNA and estrogen receptor alpha; genes associated with the epithelial-mesenchymal transition, such as β-catenin, Vimentin, and E-cadherin; and molecules associated with apoptosis. Clinical studies in bioinformatics have indicated a positive correlation between ESR1 and either CCND1 or BCL2 gene expression in all breast cancer patients. Thus, curcumin could become a potential natural adjuvant treatment for patients with estrogen receptor alpha-positive breast cancer and those with resistance or a poor response to endocrine therapy since the reactivation of estrogen receptor alpha is inevitable.
Keywords: 17ß-estradiol; antiestrogens; breast cancer; cancer therapy; curcumin; estrogen receptor alpha; estrogens.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Curcumin and epithelial-mesenchymal transition in breast cancer cells transformed by low doses of radiation and estrogen.Int J Oncol. 2016 Jun;48(6):2534-42. doi: 10.3892/ijo.2016.3477. Epub 2016 Apr 7. Int J Oncol. 2016. PMID: 27082017
-
Neo-tanshinlactone selectively inhibits the proliferation of estrogen receptor positive breast cancer cells through transcriptional down-regulation of estrogen receptor alpha.Pharmacol Res. 2016 Sep;111:849-858. doi: 10.1016/j.phrs.2016.07.044. Epub 2016 Aug 1. Pharmacol Res. 2016. PMID: 27491559
-
Keratinocyte growth factor (KGF) induces tamoxifen (Tam) resistance in human breast cancer MCF-7 cells.Anticancer Res. 2006 May-Jun;26(3A):1773-84. Anticancer Res. 2006. PMID: 16827106
-
Stabilization of MORC2 by estrogen and antiestrogens through GPER1- PRKACA-CMA pathway contributes to estrogen-induced proliferation and endocrine resistance of breast cancer cells.Autophagy. 2020 Jun;16(6):1061-1076. doi: 10.1080/15548627.2019.1659609. Epub 2019 Sep 6. Autophagy. 2020. PMID: 32401166 Free PMC article.
-
Activating Mutations of ESR1, BRCA1 and CYP19 Aromatase Genes Confer Tumor Response in Breast Cancers Treated with Antiestrogens.Recent Pat Anticancer Drug Discov. 2017;12(2):136-147. doi: 10.2174/1574892812666170227110842. Recent Pat Anticancer Drug Discov. 2017. PMID: 28245776 Review.
References
-
- WHO Breast cancer. World Health Organization. [(accessed on 20 May 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

