Analysis of the 5' Untranslated Region Length-Dependent Control of Gene Expression in Maize: A Case Study with the ZmLAZ1 Gene Family
- PMID: 39202355
- PMCID: PMC11353600
- DOI: 10.3390/genes15080994
Analysis of the 5' Untranslated Region Length-Dependent Control of Gene Expression in Maize: A Case Study with the ZmLAZ1 Gene Family
Abstract
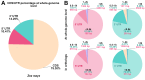
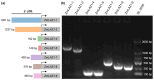
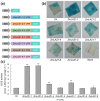
The untranslated regions (UTRs) within plant mRNAs play crucial roles in regulating gene expression and the functionality of post-translationally modified proteins by various mechanisms. These regions are vital for plants' ability to sense to multiple developmental and environmental stimuli. In this study, we conducted a genome-wide analysis of UTRs and UTR-containing genes in maize (Zea mays). Using the ZmLAZ1 family as a case study, we demonstrated that the length of 5' UTRs could influence gene expression levels by employing GUS reporter gene assays. Although maize and arabidopsis (Arabidopsis thaliana), as well as rice (Oryza sativa), have distinct functional categories of UTR-containing genes, we observed a similar lengthwise distribution of UTRs and a recurring appearance of certain gene ontology (GO) terms between maize and rice. These suggest a potentially conserved mechanism within the Poaceae species. Furthermore, the analysis of cis-acting elements in these 5' UTRs of the ZmLAZ1 gene family further supports the hypothesis that UTRs confer functional specificity to genes in a length-dependent manner. Our findings offer novel insights into the role of UTRs in maize, contributing to the broader understanding of gene expression regulation in plants.
Keywords: 5′ UTR; Lazarus 1 gene family; gene expression; maize.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
Grants and funding
- 32300318/the National Natural Science Foundation of China
- 2022M720546/the China Postdoctoral Science Foundation
- 24NSFSC1202/the Natural Science Foundation of Sichuan Province
- 2024NSFSC1324/the Natural Science Foundation of Sichuan Province
- GR-2023-E-03/the Key Laboratory of Dry-Hot Valley Characteristic Bio-Resources Development at University of Sichuan Province
LinkOut - more resources
Full Text Sources

