Interspecific and Intraspecific Transcriptomic Variations Unveil the Potential High-Altitude Adaptation Mechanisms of the Parnassius Butterfly Species
- PMID: 39202373
- PMCID: PMC11354221
- DOI: 10.3390/genes15081013
Interspecific and Intraspecific Transcriptomic Variations Unveil the Potential High-Altitude Adaptation Mechanisms of the Parnassius Butterfly Species
Abstract
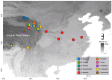
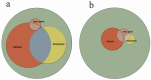
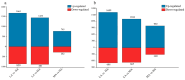
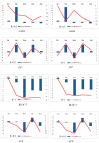
Parnassius butterflies have significantly advanced our understanding of biogeography, insect-plant interactions, and other fields of ecology and evolutionary biology. However, to date, little is known about the gene expression patterns related to the high-altitude adaptation of Parnassius species. In this study, we obtained high-throughput RNA-seq data of 48 adult Parnassius individuals covering 10 species from 12 localities in China, and deciphered their interspecific and intraspecific expression patterns based on comparative transcriptomic analyses. Though divergent transcriptional patterns among species and populations at different altitudes were found, a series of pathways related to genetic information processing (i.e., recombination, repair, transcription, RNA processing, and ribosome biogenesis), energy metabolism (i.e., oxidative phosphorylation, thermogenesis, and the citrate cycle), and cellular homeostasis were commonly enriched, reflecting similar strategies to cope with the high-altitude environments by activating energy metabolism, enhancing immune defense, and concurrently inhibiting cell growth and development. These findings deepen our understanding about the molecular mechanisms of adaptative evolution to extreme environments, and provide us with some theoretical criteria for the biodiversity conservation of alpine insects.
Keywords: Parnassius butterflies; adaptative evolution; alpine environment; high-throughput sequencing; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Su C., Ding C., Zhao Y., He B., Nie R., Hao J. Diapause-linked gene expression pattern and related candidate duplicated genes of the mountain butterfly Parnassius glacialis (Lepidoptera: Papilionidae) revealed by comprehensive transcriptome profiling. Int. J. Mol. Sci. 2023;24:5577. doi: 10.3390/ijms24065577. - DOI - PMC - PubMed
-
- Zhao J., Xia Y., Cannon C.H. Evolutionary diversification of alpine ginger reflects the early uplift of the Himalayan-Tibetan Plateau and rapid extrusion of Indochina. Gondwana Res. 2016;32:232–241. doi: 10.1016/j.gr.2015.02.004. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

