De Novo Genome Assembly of Toniniopsis dissimilis (Ramalinaceae, Lecanoromycetes) from Long Reads Shows a Comparatively High Composition of Biosynthetic Genes Putatively Involved in Melanin Synthesis
- PMID: 39202389
- PMCID: PMC11353741
- DOI: 10.3390/genes15081029
De Novo Genome Assembly of Toniniopsis dissimilis (Ramalinaceae, Lecanoromycetes) from Long Reads Shows a Comparatively High Composition of Biosynthetic Genes Putatively Involved in Melanin Synthesis
Abstract
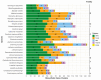
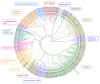
Lichens have developed numerous adaptations to optimize their survival in various environmental conditions, largely by producing secondary compounds by the fungal partner. They often have antibiotic properties and are involved in protection against intensive UV radiation, pathogens, and herbivores. To contribute to the knowledge of the arsenal of secondary compounds in a crustose lichen species, we sequenced and assembled the genome of Toniniopsis dissimilis, an indicator of old-growth forests, using Oxford Nanopore Technologies (ONT, Oxford, UK) long reads. Our analyses focused on biosynthetic gene clusters (BGCs) and specifically on Type I Polyketide (T1PKS) genes involved in the biosynthesis of polyketides. We used the comparative genomic approach to compare the genome of T. dissimilis with six other members of the family Ramalinaceae and twenty additional lichen genomes from the database. With only six T1PKS genes, a comparatively low number of biosynthetic genes are present in the T. dissimilis genome; from those, two-thirds are putatively involved in melanin biosynthesis. The comparative analyses showed at least three potential pathways of melanin biosynthesis in T. dissimilis, namely via the formation of 1,3,6,8-tetrahydroxynaphthalene, naphthopyrone, or YWA1 putative precursors, which highlights its importance in T. dissimilis. In addition, we report the occurrence of genes encoding ribosomally synthesized and posttranslationally modified peptides (RiPPs) in lichens, with their highest number in T. dissimilis compared to other Ramalinaceae genomes. So far, no function has been assigned to RiPP-like proteins in lichens, which leaves potential for future research on this topic.
Keywords: ecology; fungal RiPP-like proteins; genomics; lichen; long-read sequencing; symbiosis; type I polyketide gene.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Linking a Gene Cluster to Atranorin, a Major Cortical Substance of Lichens, through Genetic Dereplication and Heterologous Expression.mBio. 2021 Jun 29;12(3):e0111121. doi: 10.1128/mBio.01111-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154413 Free PMC article.
-
A comprehensive catalogue of polyketide synthase gene clusters in lichenizing fungi.J Ind Microbiol Biotechnol. 2018 Dec;45(12):1067-1081. doi: 10.1007/s10295-018-2080-y. Epub 2018 Sep 11. J Ind Microbiol Biotechnol. 2018. PMID: 30206732 Review.
-
Bioinformatic exploration of RiPP biosynthetic gene clusters in lichens.Fungal Biol Biotechnol. 2025 May 2;12(1):6. doi: 10.1186/s40694-025-00197-6. Fungal Biol Biotechnol. 2025. PMID: 40317021 Free PMC article.
-
Are there conserved biosynthetic genes in lichens? Genome-wide assessment of terpene biosynthetic genes suggests ubiquitous distribution of the squalene synthase cluster.BMC Genomics. 2024 Oct 7;25(1):936. doi: 10.1186/s12864-024-10806-0. BMC Genomics. 2024. PMID: 39375591 Free PMC article.
-
Recent advances in the biosynthesis of ribosomally synthesized and posttranslationally modified peptides of fungal origin.J Antibiot (Tokyo). 2023 Jan;76(1):3-13. doi: 10.1038/s41429-022-00576-w. Epub 2022 Nov 24. J Antibiot (Tokyo). 2023. PMID: 36424516 Review.
References
-
- Honegger R. 15 The Symbiotic Phenotype of Lichen-Forming Ascomycetes and Their Endo- and Epibionts. In: Hock B., editor. Fungal Associations. Springer; Berlin/Heidelberg, Germany: 2012. pp. 287–339.
-
- Schwendener S. Ueber Die Wahre Natur Der Flechten. Verh. Schweiz. Naturf. Ges. 1867;1867:88–90.
-
- Farrar J.F. The Lichen as Ecosystem: Observation and Experiment. In: Brown D.H., Hawksworth D.L., Bailey R.H., editors. Lichenology: Progress and Problems. Academic Press; London, UK: New York, NY, USA: 1976. pp. 385–406.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

