A Genotype/Phenotype Study of KDM5B-Associated Disorders Suggests a Pathogenic Effect of Dominantly Inherited Missense Variants
- PMID: 39202393
- PMCID: PMC11353349
- DOI: 10.3390/genes15081033
A Genotype/Phenotype Study of KDM5B-Associated Disorders Suggests a Pathogenic Effect of Dominantly Inherited Missense Variants
Abstract
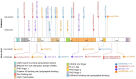
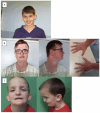
Bi-allelic disruptive variants (nonsense, frameshift, and splicing variants) in KDM5B have been identified as causative for autosomal recessive intellectual developmental disorder type 65. In contrast, dominant variants, usually disruptive as well, have been more difficult to implicate in a specific phenotype, since some of them have been found in unaffected controls or relatives. Here, we describe individuals with likely pathogenic variants in KDM5B, including eight individuals with dominant missense variants. This study is a retrospective case series of 21 individuals with variants in KDM5B. We performed deep phenotyping and collected the clinical information and molecular data of these individuals' family members. We compared the phenotypes according to variant type and to those previously described in the literature. The most common features were developmental delay, impaired intellectual development, behavioral problems, autistic behaviors, sleep disorders, facial dysmorphism, and overgrowth. DD, ASD behaviors, and sleep disorders were more common in individuals with dominant disruptive KDM5B variants, while individuals with dominant missense variants presented more frequently with renal and skin anomalies. This study extends our understanding of the KDM5B-related neurodevelopmental disorder and suggests the pathogenicity of certain dominant KDM5B missense variants.
Keywords: KDM5; epigenetics; genetic syndromes; histone demethylation; intellectual disabilities; neurodevelopmental disorders; polygenetic interactions.
Conflict of interest statement
Hans T. Bjornsson is a consultant for Mahzi Therapeutics. Paul Kruszka is an employee of GeneDx. The remaining authors declare no conflicts of interest.
Figures
References
-
- Dan J., Chen T. Chapter 3—Writers, erasers, and readers of DNA and histone methylation marks. In: Gray S.G., editor. Epigenetic Cancer Therapy. 2nd ed. Academic Press; Boston, MA, USA: 2023. pp. 39–63.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

