Molecular Manipulation of the miR160/ AUXIN RESPONSE FACTOR Expression Module Impacts Root Development in Arabidopsis thaliana
- PMID: 39202402
- PMCID: PMC11353855
- DOI: 10.3390/genes15081042
Molecular Manipulation of the miR160/ AUXIN RESPONSE FACTOR Expression Module Impacts Root Development in Arabidopsis thaliana
Abstract
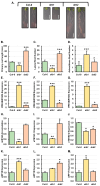
In Arabidopsis thaliana (Arabidopsis), microRNA160 (miR160) regulates the expression of AUXIN RESPONSE FACTOR10 (ARF10), ARF16 and ARF17 throughout development, including the development of the root system. We have previously shown that in addition to DOUBLE-STRANDED RNA BINDING1 (DRB1), DRB2 is also involved in controlling the rate of production of specific miRNA cohorts in the tissues where DRB2 is expressed in wild-type Arabidopsis plants. In this study, a miR160 overexpression transgene (MIR160B) and miR160-resistant transgene versions of ARF10 and ARF16 (mARF10 and mARF16) were introduced into wild-type Arabidopsis plants and the drb1 and drb2 single mutants to determine the degree of requirement of DRB2 to regulate the miR160 expression module as part of root development. Via this molecular modification approach, we show that in addition to DRB1, DRB2 is required to regulate the level of miR160 production from its precursor transcripts in Arabidopsis roots. Furthermore, we go on to correlate the altered abundance of miR160 or its ARF10, ARF16 and ARF17 target genes in the generated series of transformant lines with the enhanced development of the root system displayed by these plant lines. More specifically, promotion of primary root elongation likely stemmed from enhancement of miR160-directed ARF17 expression repression, while the promotion of lateral and adventitious root formation was the result of an elevated degree of miR160-directed regulation of ARF17 expression, and to a lesser degree, ARF10 and ARF16 expression. Taken together, the results presented in this study identify the requirement of the functional interplay between DRB1 and DRB2 to tightly control the rate of miR160 production, to in turn ensure the appropriate degree of miR160-directed ARF10, ARF16 and ARF17 gene expression regulation as part of normal root system development in Arabidopsis.
Keywords: ARF10; ARF16; ARF17; AUXIN RESPONSE FACTOR (ARF); Arabidopsis thaliana (Arabidopsis); miR160; miR160 expression module; miR160-directed gene expression regulation; microRNA (miRNA); molecular manipulation; root development.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

