A Public Mid-Density Genotyping Platform for Hexaploid Sweetpotato (Ipomoea batatas [L.] Lam)
- PMID: 39202407
- PMCID: PMC11354173
- DOI: 10.3390/genes15081047
A Public Mid-Density Genotyping Platform for Hexaploid Sweetpotato (Ipomoea batatas [L.] Lam)
Abstract
Small public breeding programs focused on specialty crops have many barriers to adopting technology, particularly creating and using genetic marker panels for genomic-based decisions in selection. Here, we report the creation of a DArTag panel of 3120 loci distributed across the sweetpotato (Ipomoea batatas [L.] Lam) genome for molecular-marker-assisted breeding and genomic prediction. The creation of this marker panel has the potential to bring cost-effective and rapid genotyping capabilities to sweetpotato breeding programs worldwide. The open access provided by this platform will allow the genetic datasets generated on the marker panel to be compared and joined across projects, institutions, and countries. This genotyping resource has the power to make routine genotyping a reality for any breeder of sweetpotato.
Keywords: DArTag genotyping; amplicon-sequencing; microhaplotype; plant breeding; sweetpotato.
Conflict of interest statement
The authors declare no conflicts of interest. There are no financial or non-financial conflicts of interest due to or resulting from collaboration with DArT, K.H.U., or DArT employes. Breeding Insight purchased services from DArT, and although the DArTag platform is proprietary to DArT, DArT will work with anyone interested in their services.
Figures



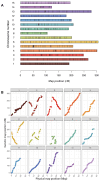
References
-
- Tanksley S.D. Molecular Markers in Plant Breeding. Plant Mol. Biol. Rep. 1983;1:3–8. doi: 10.1007/BF02680255. - DOI
-
- Feuerstein U., Brown A.H.D., Burdon J.J. Linkage of Rust Resistance Genes from Wild Barley (Hordeum Spontaneum) with Isozyme Markers. Plant Breed. 1990;104:318–324. doi: 10.1111/j.1439-0523.1990.tb00442.x. - DOI
-
- Eathington S.R., Crosbie T.M., Edwards M.D., Reiter R.S., Bull J.K. Molecular Markers in a Commercial Breeding Program. Crop Sci. 2007;47:S-154–S-163. doi: 10.2135/cropsci2007.04.0015IPBS. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

