Radiation-Tolerant Fibrivirga spp. from Rhizosphere Soil: Genome Insights and Potential in Agriculture
- PMID: 39202408
- PMCID: PMC11354047
- DOI: 10.3390/genes15081048
Radiation-Tolerant Fibrivirga spp. from Rhizosphere Soil: Genome Insights and Potential in Agriculture
Abstract
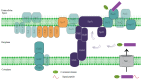
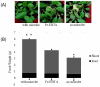
The rhizosphere of plants contains a wide range of microorganisms that can be cultivated and used for the benefit of agricultural practices. From garden soil near the rhizosphere region, Strain ES10-3-2-2 was isolated, and the cells were Gram-negative, aerobic, non-spore-forming rods that were 0.3-0.8 µm in diameter and 1.5-2.5 µm in length. The neighbor-joining method on 16S rDNA similarity revealed that the strain exhibited the highest sequence similarities with "Fibrivirga algicola JA-25" (99.2%) and Fibrella forsythia HMF5405T (97.3%). To further explore its biotechnological potentialities, we sequenced the complete genome of this strain employing the PacBio RSII sequencing platform. The genome of Strain ES10-3-2-2 comprises a 6,408,035 bp circular chromosome with a 52.8% GC content, including 5038 protein-coding genes and 52 RNA genes. The sequencing also identified three plasmids measuring 212,574 bp, 175,683 bp, and 81,564 bp. Intriguingly, annotations derived from the NCBI-PGAP, eggnog, and KEGG databases indicated the presence of genes affiliated with radiation-resistance pathway genes and plant-growth promotor key/biofertilization-related genes regarding Fe acquisition, K and P assimilation, CO2 fixation, and Fe solubilization, with essential roles in agroecosystems, as well as genes related to siderophore regulation. Additionally, T1SS, T6SS, and T9SS secretion systems are present in this species, like plant-associated bacteria. The inoculation of Strain ES10-3-2-2 to Arabidopsis significantly increases the fresh shoot and root biomass, thereby maintaining the plant quality compared to uninoculated controls. This work represents a link between radiation tolerance and the plant-growth mechanism of Strain ES10-3-2-2 based on in vitro experiments and bioinformatic approaches. Overall, the radiation-tolerant bacteria might enable the development of microbiological preparations that are extremely effective at increasing plant biomass and soil fertility, both of which are crucial for sustainable agriculture.
Keywords: Arabidopsis; PGPRs; biofertilizer; genome; siderophore.
Conflict of interest statement
The author declares no conflicts of interest.
Figures


References
-
- Olanrewaju O.S., Babalola O.O. The rhizosphere microbial complex in plant health: A review of interaction dynamics. J. Integr. Agric. 2022;21:2168–2182. doi: 10.1016/S2095-3119(21)63817-0. - DOI
-
- Kabato W.S., Janda T., Molnár Z. Unveiling the significance of rhizosphere: Implications for plant growth, stress response, and sustainable agriculture. Plant Physiol. Biochem. 2023;206:108290. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

