Transcriptional Modulation during Photomorphogenesis in Rice Seedlings
- PMID: 39202430
- PMCID: PMC11353317
- DOI: 10.3390/genes15081072
Transcriptional Modulation during Photomorphogenesis in Rice Seedlings
Erratum in
-
Correction: Gupta, P.; Jaiswal, P. Transcriptional Modulation During Photomorphogenesis in Rice Seedlings. Genes 2024, 15, 1072.Genes (Basel). 2024 Nov 27;15(12):1526. doi: 10.3390/genes15121526. Genes (Basel). 2024. PMID: 39766924 Free PMC article.
Abstract
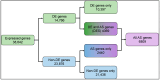
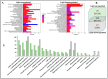
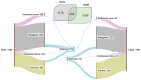
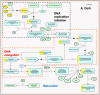
Light is one of the most important factors regulating plant gene expression patterns, metabolism, physiology, growth, and development. To explore how light may induce or alter transcript splicing, we conducted RNA-Seq-based transcriptome analyses by comparing the samples harvested as etiolated seedlings grown under continuous dark conditions vs. the light-treated green seedlings. The study aims to reveal differentially regulated protein-coding genes and novel long noncoding RNAs (lncRNAs), their light-induced alternative splicing, and their association with biological pathways. We identified 14,766 differentially expressed genes, of which 4369 genes showed alternative splicing. We observed that genes mapped to the plastid-localized methyl-erythritol-phosphate (MEP) pathway were light-upregulated compared to the cytosolic mevalonate (MVA) pathway genes. Many of these genes also undergo splicing. These pathways provide crucial metabolite precursors for the biosynthesis of secondary metabolic compounds needed for chloroplast biogenesis, the establishment of a successful photosynthetic apparatus, and photomorphogenesis. In the chromosome-wide survey of the light-induced transcriptome, we observed intron retention as the most predominant splicing event. In addition, we identified 1709 novel lncRNA transcripts in our transcriptome data. This study provides insights on light-regulated gene expression and alternative splicing in rice.
Keywords: MVA and MEP pathways; RNA-Seq; alternative splicing; circadian clock; lncRNA; photomorphogenesis; rice; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. Chapter Two—Light-Regulated Plant Growth and Development. In: Timmermans M.C.P., editor. Current Topics in Developmental Biology. Volume 91. Academic Press; Cambridge, MA, USA: 2010. pp. 29–66. Plant Development. - PubMed
-
- Naithani S., Nonogaki H., Jaiswal P. Mechanism of Plant Hormone Signaling under Stress. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2017. Exploring Crossroads Between Seed Development and Stress Response; pp. 415–454.
-
- Tobin E.M., Silverthorne J. Light Regulation of Gene Expression in Higher Plants. Annu. Rev. Plant Physiol. 1985;36:569–593. doi: 10.1146/annurev.pp.36.060185.003033. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

