Integrating ATAC-Seq and RNA-Seq Reveals the Signal Regulation Involved in the Artemia Embryonic Reactivation Process
- PMID: 39202442
- PMCID: PMC11353689
- DOI: 10.3390/genes15081083
Integrating ATAC-Seq and RNA-Seq Reveals the Signal Regulation Involved in the Artemia Embryonic Reactivation Process
Abstract
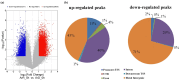
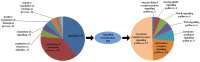
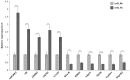
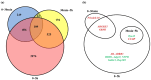
Embryonic diapause is a common evolutionary adaptation observed across a wide range of organisms. Artemia is one of the classic animal models for diapause research. The current studies of Artemia diapause mainly focus on the induction and maintenance of the embryonic diapause, with little research on the molecular regulatory mechanism of Artemia embryonic reactivation. The first 5 h after embryonic diapause breaking has been proved to be most important for embryonic reactivation in Artemia. In this work, two high-throughput sequencing methods, ATAC-seq and RNA-seq, were integrated to study the signal regulation process in embryonic reactivation of Artemia at 5 h after diapause breaking. Through the GO and KEGG enrichment analysis of the high-throughput datasets, it was showed that after 5 h of diapause breaking, the metabolism and regulation of Artemia cyst were quite active. Several signal transduction pathways were identified in the embryonic reactivation process, such as G-protein-coupled receptor (GPCR) signaling pathway, cell surface receptor signaling pathway, hormone-mediated signaling pathway, Wnt, Notch, mTOR signaling pathways, etc. It indicates that embryonic reactivation is a complex process regulated by multiple signaling pathways. With the further protein structure analysis and RT-qPCR verification, 11 GPCR genes were identified, in which 5 genes function in the embryonic reactivation stage and the other 6 genes contribute to the diapause stage. The results of this work reveal the signal transduction pathways and GPCRs involved in the embryonic reactivation process of Artemia cysts. These findings offer significant clues for in-depth research on the signal regulatory mechanisms of the embryonic reactivation process and valuable insights into the mechanism of animal embryonic diapause.
Keywords: Artemia cyst; G-protein-coupled receptor; embryonic diapause; embryonic reactivation; signal transduction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Transcriptional Regulatory Network of the Embryonic Diapause Termination Process in Artemia.Genes (Basel). 2025 Feb 1;16(2):175. doi: 10.3390/genes16020175. Genes (Basel). 2025. PMID: 40004504 Free PMC article.
-
Signaling Transduction Pathways and G-Protein-Coupled Receptors in Different Stages of the Embryonic Diapause Termination Process in Artemia.Curr Issues Mol Biol. 2024 Apr 20;46(4):3676-3693. doi: 10.3390/cimb46040229. Curr Issues Mol Biol. 2024. PMID: 38666959 Free PMC article.
-
Transcriptomic analysis elucidates the molecular processes associated with hydrogen peroxide-induced diapause termination in Artemia-encysted embryos.PLoS One. 2021 Feb 19;16(2):e0247160. doi: 10.1371/journal.pone.0247160. eCollection 2021. PLoS One. 2021. PMID: 33606769 Free PMC article.
-
Stress tolerance during diapause and quiescence of the brine shrimp, Artemia.Cell Stress Chaperones. 2016 Jan;21(1):9-18. doi: 10.1007/s12192-015-0635-7. Epub 2015 Sep 3. Cell Stress Chaperones. 2016. PMID: 26334984 Free PMC article. Review.
-
Challenges during diapause and anhydrobiosis: Mitochondrial bioenergetics and desiccation tolerance.IUBMB Life. 2018 Dec;70(12):1251-1259. doi: 10.1002/iub.1953. Epub 2018 Oct 10. IUBMB Life. 2018. PMID: 30369011 Review.
Cited by
-
Transcriptional Regulatory Network of the Embryonic Diapause Termination Process in Artemia.Genes (Basel). 2025 Feb 1;16(2):175. doi: 10.3390/genes16020175. Genes (Basel). 2025. PMID: 40004504 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

