Genome-Wide Identification of a Maize Chitinase Gene Family and the Induction of Its Expression by Fusarium verticillioides (Sacc.) Nirenberg (1976) Infection
- PMID: 39202446
- PMCID: PMC11353892
- DOI: 10.3390/genes15081087
Genome-Wide Identification of a Maize Chitinase Gene Family and the Induction of Its Expression by Fusarium verticillioides (Sacc.) Nirenberg (1976) Infection
Abstract
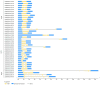
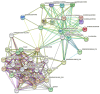
Maize chitinases are involved in chitin hydrolysis. Chitinases are distributed across various organisms including animals, plants, and fungi and are grouped into different glycosyl hydrolase families and classes, depending on protein structure. However, many chitinase functions and their interactions with other plant proteins remain unknown. The economic importance of maize (Zea mays L.) makes it relevant for studying the function of plant chitinases and their biological roles. This work aims to identify chitinase genes in the maize genome to study their gene structure, family/class classification, cis-related elements, and gene expression under biotic stress, such as Fusarium verticillioides infection. Thirty-nine chitinase genes were identified and found to be distributed in three glycosyl hydrolase (GH) families (18, 19 and 20). Likewise, the conserved domains and motifs were identified in each GH family member. The identified cis-regulatory elements are involved in plant development, hormone response, defense, and abiotic stress response. Chitinase protein-interaction network analysis predicted that they interact mainly with cell wall proteins. qRT-PCR analysis confirmed in silico data showing that ten different maize chitinase genes are induced in the presence of F. verticillioides, and that they could have several roles in pathogen infection depending on chitinase structure and cell wall localization.
Keywords: chitinase; fungi; glycosyl; hydrolase; maize; stress.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Rinaudo M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006;31:603–632. doi: 10.1016/j.progpolymsci.2006.06.001. - DOI
-
- Sahraee S., Milani J.M. Chapter 8—Chitin and chitosan-based blends, composites, and nanocomposites for packaging applications. In: Gopi S., Thomas S., Pius A., editors. Handbook of Chitin and Chitosan. Elsevier; Amsterdam, The Netherlands: 2020. pp. 247–271.
-
- Aranaz I., Mengibar M., Harris R., Panos I., Miralles B., Acosta N., Galed G., Heras A. Functional characterization of chitin and chitosan. Curr. Chem. Biol. 2009;3:203–230. doi: 10.2174/187231309788166415. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

