In Silico Analysis and Development of the Secretory Expression of D-Psicose-3-Epimerase in Escherichia coli
- PMID: 39203416
- PMCID: PMC11356227
- DOI: 10.3390/microorganisms12081574
In Silico Analysis and Development of the Secretory Expression of D-Psicose-3-Epimerase in Escherichia coli
Abstract
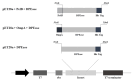
D-psicose-3-epimerase (DPEase), a key enzyme for D-psicose production, has been successfully expressed in Escherichia coli with high yield. However, intracellular expression results in high downstream processing costs and greater risk of lipopolysaccharide (LPS) contamination during cell disruption. The secretory expression of DPEase could minimize the number of purification steps and prevent LPS contamination, but achieving the secretion expression of DPEase in E. coli is challenging and has not been reported due to certain limitations. This study addresses these challenges by enhancing the secretion of DPEase in E. coli through computational predictions and structural analyses. Signal peptide prediction identified PelB as the most effective signal peptide for DPEase localization and enhanced solubility. Supplementary strategies included the addition of 0.1% (v/v) Triton X-100 to promote protein secretion, resulting in higher extracellular DPEase (0.5 unit/mL). Low-temperature expression (20 °C) mitigated the formation of inclusion bodies, thus enhancing DPEase solubility. Our findings highlight the pivotal role of signal peptide selection in modulating DPEase solubility and activity, offering valuable insights for protein expression and secretion studies, especially for rare sugar production. Ongoing exploration of alternative signal peptides and refinement of secretion strategies promise further enhancement in enzyme secretion efficiency and process safety, paving the way for broader applications in biotechnology.
Keywords: D-psicose-3-epimerase; protein expression; protein secretion; psicose; signal peptide.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Patra F., Patel A., Shah N. Microbial Production of Low-Calorie Sugars. In: Holban A.M., Grumezescu A.M., editors. Handbook of Food Bioengineering, Microbial Production of Food Ingredients and Additives. 1st ed. Volume 5. Academic Press; London, UK: 2017. pp. 259–290. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

