A Bead Biofilm Reactor for High-Throughput Growth and Translational Applications
- PMID: 39203430
- PMCID: PMC11356137
- DOI: 10.3390/microorganisms12081588
A Bead Biofilm Reactor for High-Throughput Growth and Translational Applications
Abstract
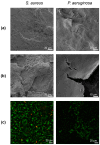
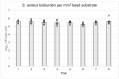
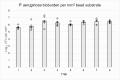
Bacteria in natural ecosystems such as soil, dirt, or debris preferentially reside in the biofilm phenotype. When a traumatic injury, such as an open fracture, occurs, these naturally dwelling biofilms and accompanying foreign material can contaminate the injury site. Given their high tolerance of systemic levels of antibiotics that may be administered prophylactically, biofilms may contribute to difficult-to-treat infections. In most animal models, planktonic bacteria are used as initial inocula to cause infection, and this might not accurately mimic clinically relevant contamination and infection scenarios. Further, few approaches and systems utilize the same biofilm and accompanying substrate throughout the experimental continuum. In this study, we designed a unique reactor to grow bacterial biofilms on up to 50 silica beads that modeled environmental wound contaminants. The data obtained indicated that the reactor system repeatably produced mature Staphylococcus aureus and Pseudomonas aeruginosa biofilms on the silica beads, with an average of 5.53 and 6.21 log10 colony-forming units per mm2, respectively. The bead substrates are easily manipulable for in vitro or in vivo applications, thus improving translatability. Taken together, the bead biofilm reactor presented herein may be a useful system for repeatably growing established biofilms on silica beads that could be used for susceptibility testing and as initial inocula in future animal models of trauma-related injuries.
Keywords: P. aeruginosa; S. aureus; SEM; antibiofilm; biofilm; in vitro; reactor; repeatability; surface coverage.
Conflict of interest statement
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the United States Government.
Figures


Similar articles
-
Biofilm Growth on Simulated Fracture Fixation Plates Using a Customized CDC Biofilm Reactor for a Sheep Model of Biofilm-Related Infection.Microorganisms. 2022 Mar 31;10(4):759. doi: 10.3390/microorganisms10040759. Microorganisms. 2022. PMID: 35456808 Free PMC article.
-
A modified CDC biofilm reactor to produce mature biofilms on the surface of peek membranes for an in vivo animal model application.Curr Microbiol. 2011 Jun;62(6):1657-63. doi: 10.1007/s00284-011-9908-2. Epub 2011 Mar 25. Curr Microbiol. 2011. PMID: 21437591
-
Halicin Is Effective Against Staphylococcus aureus Biofilms In Vitro.Clin Orthop Relat Res. 2022 Aug 1;480(8):1476-1487. doi: 10.1097/CORR.0000000000002251. Epub 2022 May 17. Clin Orthop Relat Res. 2022. PMID: 35583504 Free PMC article.
-
Effectiveness of a polyhexamethylene biguanide-containing wound cleansing solution using experimental biofilm models.J Wound Care. 2023 Jun 2;32(6):359-367. doi: 10.12968/jowc.2023.32.6.359. J Wound Care. 2023. PMID: 37300862
-
In vitro testing of a first-in-class tri-alkylnorspermidine-biaryl antibiotic in an anti-biofilm silicone coating.Acta Biomater. 2019 Jul 15;93:25-35. doi: 10.1016/j.actbio.2019.02.010. Epub 2019 Feb 12. Acta Biomater. 2019. PMID: 30769135
Cited by
-
In vitro antibiofilm efficacy of ertapenem, tobramycin, and moxifloxacin against biofilms grown in a glass bead or CDC Biofilm Reactor®.PLoS One. 2025 Feb 10;20(2):e0318487. doi: 10.1371/journal.pone.0318487. eCollection 2025. PLoS One. 2025. PMID: 39928650 Free PMC article.
References
-
- Lewis K. Persister cells: Molecular mechanisms related to antibiotic tolerance. Antibiot. Resist. 2012;211:121–133. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

