Resistance and Co-Resistance of Metallo-Beta-Lactamase Genes in Diarrheal and Urinary-Tract Pathogens in Bangladesh
- PMID: 39203431
- PMCID: PMC11356267
- DOI: 10.3390/microorganisms12081589
Resistance and Co-Resistance of Metallo-Beta-Lactamase Genes in Diarrheal and Urinary-Tract Pathogens in Bangladesh
Abstract
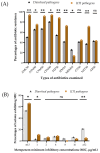
Carbapenems are the antibiotics of choice for treating multidrug-resistant bacterial infections. Metallo-β-lactamases (MBLs) are carbapenemases capable of hydrolyzing nearly all therapeutically available beta-lactam antibiotics. Consequently, this research assessed the distribution of two MBL genes and three β-lactamases and their associated phenotypic resistance in diarrheal and urinary-tract infections (UTIs) to guide future policies. Samples were collected through a cross-sectional study, and β-lactamase genes were detected via PCR. A total of 228 diarrheal bacteria were isolated from 240 samples. The most predominant pathogens were Escherichia coli (32%) and Klebsiella spp. (7%). Phenotypic resistance to amoxicillin-clavulanic acid, aztreonam, cefuroxime, cefixime, cefepime, imipenem, meropenem, gentamicin, netilmicin, and amikacin was 50.4%, 65.6%, 66.8%, 80.5%, 54.4%, 41.6%, 25.7%, 41.2%, 37.2%, and 42.9%, respectively. A total of 142 UTI pathogens were identified from 150 urine samples. Klebsiella spp. (39%) and Escherichia coli (24%) were the major pathogens isolated. Phenotypic resistance to amoxicillin-clavulanic acid, aztreonam, cefuroxime, cefixime, cefepime, imipenem, meropenem, gentamicin, netilmicin, and amikacin was 93.7%, 75.0%, 91.5%, 93.7%, 88.0%, 72.5%, 13.6%, 44.4%, 71.1%, and 43%, respectively. Twenty-four diarrheal isolates carried blaNDM-1 or blaVIM genes. The overall MBL gene prevalence was 10.5%. Thirty-six UTI pathogens carried either blaNDM-1 or blaVIM genes (25.4%). Seven isolates carried both blaNDM-1 and blaVIM genes. MBL genes were strongly associated with phenotypic carbapenem and other β-lactam antibiotic resistance. blaOXA imparted significantly higher phenotypic resistance to β-lactam antibiotics. Active surveillance and stewardship programs are urgently needed to reduce carbapenem resistance in Bangladesh.
Keywords: Bangladesh; antimicrobial resistance; antimicrobial stewardship programs; blaNDM-1; blaVIM; co-resistance; diarrhea; metallo-beta-lactamase; urinary tract infections.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Genotypic to Phenotypic Resistance Discrepancies Identified Involving β-Lactamase Genes, blaKPC, blaIMP, blaNDM-1, and blaVIM in Uropathogenic Klebsiella pneumoniae.Infect Drug Resist. 2020 Aug 18;13:2863-2875. doi: 10.2147/IDR.S262493. eCollection 2020. Infect Drug Resist. 2020. PMID: 32903880 Free PMC article.
-
Prevalence of Metallo-β-lactamase Producing Non-fermentative Pseudomonas Species from Clinical Isolates in Dhaka, Bangladesh.Mymensingh Med J. 2018 Jan;27(1):89-94. Mymensingh Med J. 2018. PMID: 29459597
-
[Molecular Analysis of Antibiotic Resistance in CarbapenemResistant Pseudomonas aeruginosa Isolates Isolated from Clinical Samples].Mikrobiyol Bul. 2024 Apr;58(2):148-170. doi: 10.5578/mb.202498200. Mikrobiyol Bul. 2024. PMID: 38676583 Turkish.
-
Sulopenem: An Intravenous and Oral Penem for the Treatment of Urinary Tract Infections Due to Multidrug-Resistant Bacteria.Drugs. 2022 Apr;82(5):533-557. doi: 10.1007/s40265-022-01688-1. Epub 2022 Mar 16. Drugs. 2022. PMID: 35294769 Review.
-
Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli.Drugs. 2019 Feb;79(3):271-289. doi: 10.1007/s40265-019-1055-2. Drugs. 2019. PMID: 30712199 Review.
Cited by
-
Editorial: New therapeutic strategies against carbapenem-resistant gram-negative bacteria.Front Microbiol. 2024 Nov 7;15:1513900. doi: 10.3389/fmicb.2024.1513900. eCollection 2024. Front Microbiol. 2024. PMID: 39575187 Free PMC article. No abstract available.
References
-
- Sharland M., Zanichelli V., Ombajo L.A., Bazira J., Cappello B., Chitatanga R., Chuki P., Gandra S., Getahun H., Harbarth S. The WHO essential medicines list AWaRe book: From a list to a quality improvement system. Clin. Microbiol. Infect. 2022;28:1533–1535. doi: 10.1016/j.cmi.2022.08.009. - DOI - PubMed
-
- Sharland M., Gandra S., Huttner B., Moja L., Pulcini C., Zeng M., Mendelson M., Cappello B., Cooke G., Magrini N. Encouraging AWaRe-ness and discouraging inappropriate antibiotic use—The new 2019 Essential Medicines List becomes a global antibiotic stewardship tool. Lancet Infect. Dis. 2019;19:1278–1280. doi: 10.1016/S1473-3099(19)30532-8. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

