Molecular Evolutionary Analyses of the Fusion Genes in Human Parainfluenza Virus Type 4
- PMID: 39203475
- PMCID: PMC11356533
- DOI: 10.3390/microorganisms12081633
Molecular Evolutionary Analyses of the Fusion Genes in Human Parainfluenza Virus Type 4
Abstract
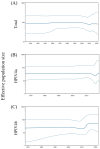
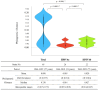
The human parainfluenza virus type 4 (HPIV4) can be classified into two distinct subtypes, 4a and 4b. The full lengths of the fusion gene (F gene) of 48 HPIV4 strains collected during the period of 1966-2022 were analyzed. Based on these gene sequences, the time-scaled evolutionary tree was constructed using Bayesian Markov chain Monte Carlo methods. A phylogenetic tree showed that the first division of the two subtypes occurred around 1823, and the most recent common ancestors of each type, 4a and 4b, existed until about 1940 and 1939, respectively. Although the mean genetic distances of all strains were relatively wide, the distances in each subtype were not wide, indicating that this gene was conserved in each subtype. The evolutionary rates of the genes were relatively low (4.41 × 10-4 substitutions/site/year). Moreover, conformational B-cell epitopes were predicted in the apex of the trimer fusion protein. These results suggest that HPIV4 subtypes diverged 200 years ago and the progenies further diverged and evolved.
Keywords: fusion gene; human parainfluenza virus 4; molecular evolution.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Kimura H., Hayashi Y., Kitagawa M., Yoshizaki M., Saito K., Harada K., Okayama K., Miura Y., Kimura R., Shirai T., et al. Pathogen Profiles in Outpatients with Non-COVID-19 during the 7th Prevalent Period of COVID-19 in Gunma, Japan. Microorganisms. 2023;11:2142. doi: 10.3390/microorganisms11092142. - DOI - PMC - PubMed
-
- Wang X., Li Y., Deloria-Knoll M., Madhi S.A., Cohen C., Arguelles V.L., Basnet S., Bassat Q., Brooks W.A., Echavarria M., et al. Global burden of acute lower respiratory infection associated with human parainfluenza virus in children younger than 5 years for 2018: A systematic review and meta-analysis. Lancet Glob. Health. 2021;9:e1077–e1087. doi: 10.1016/S2214-109X(21)00218-7. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

