Genomic Analysis of a Novel Torradovirus "Rehmannia Torradovirus Virus": Two Distinct Variants Infecting Rehmannia glutinosa
- PMID: 39203485
- PMCID: PMC11356386
- DOI: 10.3390/microorganisms12081643
Genomic Analysis of a Novel Torradovirus "Rehmannia Torradovirus Virus": Two Distinct Variants Infecting Rehmannia glutinosa
Abstract
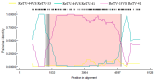
Rehmannia glutinosa, a crucial medicinal plant native to China, is extensively cultivated across East Asia. We used high-throughput sequencing to identify viruses infecting R. glutinosa with mosaic, leaf yellowing, and necrotic symptoms. A novel Torradovirus, which we tentatively named "Rehmannia torradovirus virus" (ReTV), was identified. The complete sequences were obtained through reverse-transcription polymerase chain reaction (RT-PCR), 5' and 3' rapid amplification of cDNA ends, and Sanger sequencing. The amino acid sequence alignment between the ReTV-52 isolate and known Torradovirus species in the Pro-Pol and coat protein regions were 51.3-73.3% and 37.1-68.1%, respectively. Meanwhile, the amino acid sequence alignment between the ReTV-8 isolate and known Torradovirus species in the Pro-Pol and coat protein regions were 52.7-72.8% and 36.8-67.5%, respectively. The sequence analysis classified ten ReTV strains into two variants. The ReTV-52 genome has two RNA segments of 6939 and 4569 nucleotides, while that of ReTV-8 consists of two RNA segments containing 6889 and 4662 nucleotides. Sequence comparisons and phylogenetic analysis showed ReTV strains clustered within the Torradovirus, exhibiting the closet relation to the squash chlorotic leaf spot virus. The RT-PCR results showed a 100% ReTV detection rate in all 60 R. glutinosa samples. Therefore, ReTV should be classified as a novel Torradovirus species. ReTV is potentially dangerous to R. glutinosa, and necessitating monitoring this virus in the field.
Keywords: Rehmannia glutinosa; Rehmannia torradovirus virus; Torradoviruses; high-throughput sequencing; phylogenetic relationship; sequence comparison.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Kwak H.R., Go W.R., Hong S.B., Kim J.E., Kim M., Choi H.S. One-step multiplex RT-PCR for simultaneous detection of four viruses infecting Rehmannia glutinosa. J. Gen. Plant Pathol. 2020;86:143–148. doi: 10.1007/s10327-019-00904-3. - DOI
-
- Zhi J.Y., Li Y.J., Zhang Z.Y., Yang C.F., Geng X.T., Zhang M., Li X.R., Zuo X., Li M.J., Huang Y., et al. Molecular regulation of catalpol and acteoside accumulation in radial striation and non-radial striation of Rehmannia glutinosa tuberous root. Int. J. Mol. Sci. 2018;19:3751. doi: 10.3390/ijms19123751. - DOI - PMC - PubMed
-
- Liu D., Dong C.M. Discussion on the origin change of Rehmannia glutinosa in Henan Province. Heilongjiang Agric. Sci. 2018;4:133–136.
-
- Ling H., Liu R.R. Progress in medicinal plant Rehmannia glutinosa: Metabolite profiling, tissue culture, growth and its regulation, and functional genomics. Russ. J. Plant Physiol. 2009;56:591–598. doi: 10.1134/S1021443709050021. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

