Novel Competitive ELISA Utilizing Trimeric Spike Protein of SARS-CoV-2, Could Identify More Than RBD-RBM Specific Neutralizing Antibodies in Hybrid Sera
- PMID: 39204038
- PMCID: PMC11359269
- DOI: 10.3390/vaccines12080914
Novel Competitive ELISA Utilizing Trimeric Spike Protein of SARS-CoV-2, Could Identify More Than RBD-RBM Specific Neutralizing Antibodies in Hybrid Sera
Abstract
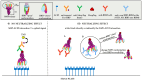
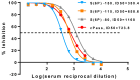
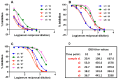
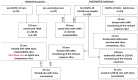
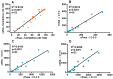
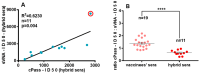
Since the initiation of the COVID-19 pandemic, there has been a need for the development of diagnostic methods to determine the factors implicated in mounting an immune response against the virus. The most promising indicator has been suggested to be neutralizing antibodies (nAbs), which mainly block the interaction between the Spike protein (S) of SARS-CoV-2 and the host entry receptor ACE2. In this study, we aimed to develop and optimize conditions of a competitive ELISA to measure serum neutralizing titer, using a recombinant trimeric Spike protein modified to have six additional proline residues (S(6P)-HexaPro) and h-ACE2. The results of our surrogate Virus Neutralizing Assay (sVNA) were compared against the commercial sVNT (cPass, Nanjing GenScript Biotech Co., Nanjing City, China), using serially diluted sera from vaccinees, and a high correlation of ID50-90 titer values was observed between the two assays. Interestingly, when we tested and compared the neutralizing activity of sera from eleven fully vaccinated individuals who subsequently contracted COVID-19 (hybrid sera), we recorded a moderate correlation between the two assays, while higher sera neutralizing titers were measured with sVNA. Our data indicated that the sVNA, as a more biologically relevant model assay that paired the trimeric S(6P) with ACE2, instead of the isolated RBD-ACE2 pairing cPass test, could identify nAbs other than the RBD-RBM specific ones.
Keywords: SARS-CoV-2; non-RBD-RBM neutralizing antibodies (nAbs); surrogate viral neutralization assay (sVNA); trimeric S(6P)-HexaPro.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
A highly sensitive bead-based flow cytometric competitive binding assay to detect SARS-CoV-2 neutralizing antibody activity.Front Immunol. 2022 Nov 30;13:1041860. doi: 10.3389/fimmu.2022.1041860. eCollection 2022. Front Immunol. 2022. PMID: 36532082 Free PMC article.
-
Evaluation of a biotin-based surrogate virus neutralization test for detecting postvaccination antibodies against SARS-CoV-2 variants in sera.Biochem Biophys Res Commun. 2023 Feb 26;646:8-18. doi: 10.1016/j.bbrc.2023.01.052. Epub 2023 Jan 19. Biochem Biophys Res Commun. 2023. PMID: 36696754 Free PMC article.
-
Evaluation of commercially available fully automated and ELISA-based assays for detecting anti-SARS-CoV-2 neutralizing antibodies.Sci Rep. 2022 Nov 8;12(1):19020. doi: 10.1038/s41598-022-21317-x. Sci Rep. 2022. PMID: 36347859 Free PMC article.
-
Competitive SARS-CoV-2 Serology Reveals Most Antibodies Targeting the Spike Receptor-Binding Domain Compete for ACE2 Binding.mSphere. 2020 Sep 16;5(5):e00802-20. doi: 10.1128/mSphere.00802-20. mSphere. 2020. PMID: 32938700 Free PMC article.
-
A Combination of Receptor-Binding Domain and N-Terminal Domain Neutralizing Antibodies Limits the Generation of SARS-CoV-2 Spike Neutralization-Escape Mutants.mBio. 2021 Oct 26;12(5):e0247321. doi: 10.1128/mBio.02473-21. Epub 2021 Oct 5. mBio. 2021. PMID: 34607456 Free PMC article.
Cited by
-
Advances in Surrogate Neutralization Tests for High-Throughput Screening and the Point-of-Care.Anal Chem. 2025 Mar 18;97(10):5407-5423. doi: 10.1021/acs.analchem.5c00666. Epub 2025 Mar 4. Anal Chem. 2025. PMID: 40035475 Free PMC article. Review. No abstract available.
-
Nanoparticle-supported, rapid, digital quantification of neutralizing antibodies against SARS-CoV-2 variants.Biosens Bioelectron. 2025 Oct 1;285:117549. doi: 10.1016/j.bios.2025.117549. Epub 2025 May 7. Biosens Bioelectron. 2025. PMID: 40383030
References
LinkOut - more resources
Full Text Sources
Miscellaneous

