Semi-Covariance Coefficient Analysis of Spike Proteins from SARS-CoV-2 and Its Variants Omicron, BA.5, EG.5, and JN.1 for Viral Infectivity, Virulence and Immune Escape
- PMID: 39205166
- PMCID: PMC11360586
- DOI: 10.3390/v16081192
Semi-Covariance Coefficient Analysis of Spike Proteins from SARS-CoV-2 and Its Variants Omicron, BA.5, EG.5, and JN.1 for Viral Infectivity, Virulence and Immune Escape
Abstract
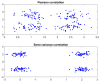
Semi-covariance has attracted significant attention in recent years and is increasingly employed to elucidate statistical phenomena exhibiting fluctuations, such as the similarity or difference in charge patterns of spike proteins among coronaviruses. In this study, by examining values above and below the average/mean based on the positive and negative charge patterns of amino acid residues in the spike proteins of SARS-CoV-2 and its current circulating variants, the proposed methods offer profound insights into the nonlinear evolving trends in those viral spike proteins. Our study indicates that the charge span value can predict the infectivity of the virus and the charge density can estimate the virulence of the virus, and both predicated infectivity and virulence appear to be associated with the capability of viral immune escape. This semi-covariance coefficient analysis may be used not only to predict the infectivity, virulence and capability of immune escape for coronaviruses but also to analyze the functionality of other viral proteins. This study improves our understanding of the trend of viral evolution in terms of viral infectivity, virulence or the capability of immune escape, which remains further validated by more future studies and statistical data.
Keywords: SARS-CoV-2; coronaviruses; immune escape; infectivity; semi-covariance coefficient; spike protein sequence; variants; virulence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Peters E.E. Fractal DNA Analysis: Applying Chaos Theory to Investment and Economics. Wiley; Hoboken, NJ, USA: 1994.
-
- Buldyrev S.V., Goldberger A.L., Havlin S., Mantegna R.N., Matsa M.E., Peng C.K., Simons M., Stanley H.E. Long-range correlation properties of coding and noncoding DNA sequences: GenBank analysis. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 1995;51:5084–5091. doi: 10.1103/PhysRevE.51.5084. - DOI - PubMed
-
- Xu Q., Bai Y. Semiparametric statistical inferences for longitudinal data with nonparametric covariance modelling. Statistics. 2017;51:1280–1303. doi: 10.1080/02331888.2017.1354861. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

