Interaction between SARS-CoV PBM and Cellular PDZ Domains Leading to Virus Virulence
- PMID: 39205188
- PMCID: PMC11359647
- DOI: 10.3390/v16081214
Interaction between SARS-CoV PBM and Cellular PDZ Domains Leading to Virus Virulence
Abstract
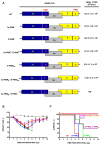
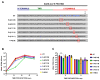
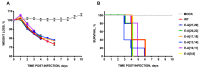
The interaction between SARS-CoV PDZ-binding motifs (PBMs) and cellular PDZs is responsible for virus virulence. The PBM sequence present in the 3a and envelope (E) proteins of SARS-CoV can potentially bind to over 400 cellular proteins containing PDZ domains. The role of SARS-CoV 3a and E proteins was studied. SARS-CoVs, in which 3a-PBM and E-PMB have been deleted (3a-PBM-/E-PBM-), reduced their titer around one logarithmic unit but still were viable. In addition, the absence of the E-PBM and the replacement of 3a-PBM with that of E did not allow the rescue of SARS-CoV. E protein PBM was necessary for virulence, activating p38-MAPK through the interaction with Syntenin-1 PDZ domain. However, the presence or absence of the homologous motif in the 3a protein, which does not bind to Syntenin-1, did not affect virus pathogenicity. Mutagenesis analysis and in silico modeling were performed to study the extension of the PBM of the SARS-CoV E protein. Alanine and glycine scanning was performed revealing a pair of amino acids necessary for optimum virus replication. The binding of E protein with the PDZ2 domain of the Syntenin-1 homodimer induced conformational changes in both PDZ domains 1 and 2 of the dimer.
Keywords: PBM-PDZ; coronavirus; virulence; virus–host interaction.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

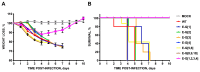


References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.I., Lauber C., Leontovich A.M., Neuman B.W., et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. - PMC - PubMed
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
-
- Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., Sola I., Enjuanes L. Viral PDZ Binding Motifs Influence Cell Behavior Through the Interaction with Cellular Proteins Containing PDZ Domains. Methods Mol. Biol. 2021;2256:217–236. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

