An ISG15-Based High-Throughput Screening Assay for Identification and Characterization of SARS-CoV-2 Inhibitors Targeting Papain-like Protease
- PMID: 39205213
- PMCID: PMC11359932
- DOI: 10.3390/v16081239
An ISG15-Based High-Throughput Screening Assay for Identification and Characterization of SARS-CoV-2 Inhibitors Targeting Papain-like Protease
Abstract
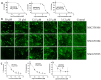
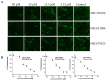
Emergence of newer variants of SARS-CoV-2 underscores the need for effective antivirals to complement the vaccination program in managing COVID-19. The multi-functional papain-like protease (PLpro) of SARS-CoV-2 is an essential viral protein that not only regulates the viral replication but also modulates the host immune system, making it a promising therapeutic target. To this end, we developed an in vitro interferon stimulating gene 15 (ISG15)-based Förster resonance energy transfer (FRET) assay and screened the National Cancer Institute (NCI) Diversity Set VI compound library, which comprises 1584 small molecules. Subsequently, we assessed the PLpro enzymatic activity in the presence of screened molecules. We identified three potential PLpro inhibitors, namely, NSC338106, 651084, and 679525, with IC50 values in the range from 3.3 to 6.0 µM. These molecules demonstrated in vitro inhibition of the enzyme activity and exhibited antiviral activity against SARS-CoV-2, with EC50 values ranging from 0.4 to 4.6 µM. The molecular docking of all three small molecules to PLpro suggested their specificity towards the enzyme's active site. Overall, our study contributes promising prospects for further developing potential antivirals to combat SARS-CoV-2 infection.
Keywords: FRET assay; ISG15; PLpro; SARS-CoV-2; high-throughput screening.
Conflict of interest statement
A patent has been filed for this study. The authors declare no conflicts of interest.
Figures





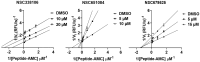

References
-
- Puhach O., Adea K., Hulo N., Sattonnet P., Genecand C., Iten A., Jacquérioz F., Kaiser L., Vetter P., Eckerle I., et al. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS-CoV-2. Nat. Med. 2022;28:1491–1500. doi: 10.1038/s41591-022-01816-0. - DOI - PubMed
-
- Ong S.W.X., Chiew C.J., Ang L.W., Mak T.M., Cui L., Toh M., Lim Y.D., Lee P.H., Lee T.H., Chia P.Y., et al. Clinical and Virological Features of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants of Concern: A Retrospective Cohort Study Comparing B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta) Clin. Infect. Dis. 2022;75:e1128–e1136. doi: 10.1093/cid/ciab721. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

