Molecular Bases and Specificity behind the Activation of the Immune System OAS/RNAse L Pathway by Viral RNA
- PMID: 39205220
- PMCID: PMC11359028
- DOI: 10.3390/v16081246
Molecular Bases and Specificity behind the Activation of the Immune System OAS/RNAse L Pathway by Viral RNA
Abstract
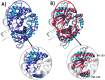
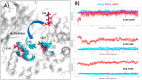
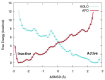
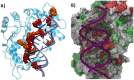
The first line of defense against invading pathogens usually relies on innate immune systems. In this context, the recognition of exogenous RNA structures is primordial to fight, notably, against RNA viruses. One of the most efficient immune response pathways is based on the sensing of RNA double helical motifs by the oligoadenylate synthase (OAS) proteins, which in turn triggers the activity of RNase L and, thus, cleaves cellular and viral RNA. In this contribution, by using long-range molecular dynamics simulations, complemented with enhanced sampling techniques, we elucidate the structural features leading to the activation of OAS by interaction with a model double-strand RNA oligomer mimicking a viral RNA. We characterize the allosteric regulation induced by the nucleic acid leading to the population of the active form of the protein. Furthermore, we also identify the free energy profile connected to the active vs. inactive conformational transitions in the presence and absence of RNA. Finally, the role of two RNA mutations, identified as able to downregulate OAS activation, in shaping the protein/nucleic acid interface and the conformational landscape of OAS is also analyzed.
Keywords: RNA viruses; free energy profiles; innate immune system; molecular dynamics; oligoadenylate synthase.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
New insights into the role of RNase L in innate immunity.J Interferon Cytokine Res. 2011 Jan;31(1):49-57. doi: 10.1089/jir.2010.0120. Epub 2010 Dec 29. J Interferon Cytokine Res. 2011. PMID: 21190483 Free PMC article. Review.
-
Role of helical structure and dynamics in oligoadenylate synthetase 1 (OAS1) mismatch tolerance and activation by short dsRNAs.Proc Natl Acad Sci U S A. 2022 Jan 18;119(3):e2107111119. doi: 10.1073/pnas.2107111119. Proc Natl Acad Sci U S A. 2022. PMID: 35017296 Free PMC article.
-
The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities.J Interferon Cytokine Res. 2011 Jan;31(1):41-7. doi: 10.1089/jir.2010.0107. Epub 2010 Dec 12. J Interferon Cytokine Res. 2011. PMID: 21142819 Review.
-
Rotavirus Controls Activation of the 2'-5'-Oligoadenylate Synthetase/RNase L Pathway Using at Least Two Distinct Mechanisms.J Virol. 2015 Dec;89(23):12145-53. doi: 10.1128/JVI.01874-15. Epub 2015 Sep 23. J Virol. 2015. PMID: 26401041 Free PMC article.
-
A human cellular noncoding RNA activates the antiviral protein 2'-5'-oligoadenylate synthetase 1.J Biol Chem. 2018 Oct 12;293(41):16115-16124. doi: 10.1074/jbc.RA118.004747. Epub 2018 Aug 20. J Biol Chem. 2018. PMID: 30126839 Free PMC article.
References
-
- Chakrabartty I., Khan M., Mahanta S., Chopra H., Dhawan M., Choudhary O.P., Bibi S., Mohanta Y.K., Emran T. Bin Comparative overview of emerging RNA viruses: Epidemiology, pathogenesis, diagnosis and current treatment. Ann. Med. Surg. 2022;79:103985. doi: 10.1016/j.amsu.2022.103985. - DOI - PMC - PubMed
-
- Tchicaya A., Lorentz N., Leduc K., de Lanchy G. COVID-19 mortality with regard to healthcare services availability, health risks, and socio-spatial factors at department level in France: A spatial cross-sectional analysis. PLoS ONE. 2021;16:e0256857. doi: 10.1371/journal.pone.0256857. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

