A Bioinformatic Ecosystem for Bacteriophage Genomics: PhaMMSeqs, Phamerator, pdm_utils, PhagesDB, DEPhT, and PhamClust
- PMID: 39205252
- PMCID: PMC11359507
- DOI: 10.3390/v16081278
A Bioinformatic Ecosystem for Bacteriophage Genomics: PhaMMSeqs, Phamerator, pdm_utils, PhagesDB, DEPhT, and PhamClust
Abstract
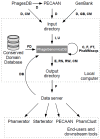
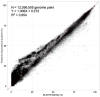
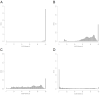
The last thirty years have seen a meteoric rise in the number of sequenced bacteriophage genomes, spurred on by both the rise and success of groups working to isolate and characterize phages, and the rapid and significant technological improvements and reduced costs associated with sequencing their genomes. Over the course of these decades, the tools used to glean evolutionary insights from these sequences have grown more complex and sophisticated, and we describe here the suite of computational and bioinformatic tools used extensively by the integrated research-education communities such as SEA-PHAGES and PHIRE, which are jointly responsible for 25% of all complete phage genomes in the RefSeq database. These tools are used to integrate and analyze phage genome data from different sources, for identification and precise extraction of prophages from bacterial genomes, computing "phamilies" of related genes, and displaying the complex nucleotide and amino acid level mosaicism of these genomes. While over 50,000 SEA-PHAGES students have primarily benefitted from these tools, they are freely available for the phage community at large.
Keywords: Actinobacteria; Actinobacteriophage; Bacteriophage; genomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

