The Disorderly Nature of Caliciviruses
- PMID: 39205298
- PMCID: PMC11360831
- DOI: 10.3390/v16081324
The Disorderly Nature of Caliciviruses
Abstract
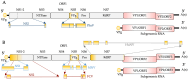
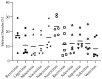
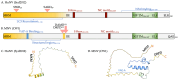
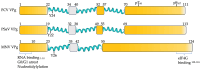
An intrinsically disordered protein (IDP) or region (IDR) lacks or has little protein structure but still maintains function. This lack of structure creates flexibility and fluidity, allowing multiple protein conformations and potentially transient interactions with more than one partner. Caliciviruses are positive-sense ssRNA viruses, containing a relatively small genome of 7.6-8.6 kb and have a broad host range. Many viral proteins are known to contain IDRs, which benefit smaller viral genomes by expanding the functional proteome through the multifunctional nature of the IDR. The percentage of intrinsically disordered residues within the total proteome for each calicivirus type species can range between 8 and 23%, and IDRs have been experimentally identified in NS1-2, VPg and RdRP proteins. The IDRs within a protein are not well conserved across the genera, and whether this correlates to different activities or increased tolerance to mutations, driving virus adaptation to new selection pressures, is unknown. The function of norovirus NS1-2 has not yet been fully elucidated but includes involvement in host cell tropism, the promotion of viral spread and the suppression of host interferon-λ responses. These functions and the presence of host cell-like linear motifs that interact with host cell caspases and VAPA/B are all found or affected by the disordered region of norovirus NS1-2. The IDRs of calicivirus VPg are involved in viral transcription and translation, RNA binding, nucleotidylylation and cell cycle arrest, and the N-terminal IDR within the human norovirus RdRP could potentially drive liquid-liquid phase separation. This review identifies and summarises the IDRs of proteins within the Caliciviridae family and their importance during viral replication and subsequent host interactions.
Keywords: NS1-2; RdRP; VPg; calicivirus; intrinsically disordered protein; norovirus; sapovirus; vesivirus.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- Mantonico M.V., De Leo F., Quilici G., Colley L.S., De Marchis F., Crippa M., Mezzapelle R., Schulte T., Zucchelli C., Pastorello C., et al. The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12. Nat. Commun. 2024;15:1201. doi: 10.1038/s41467-024-45505-7. - DOI - PMC - PubMed
-
- Dominguez F., Shiliaev N., Lukash T., Agback P., Palchevska O., Gould J.R., Meshram C.D., Prevelige P.E., Green T.J., Agback T., et al. NAP1L1 and NAP1L4 Binding to Hypervariable Domain of Chikungunya Virus nsP3 Protein Is Bivalent and Requires Phosphorylation. J. Virol. 2021;95:e0083621. doi: 10.1128/JVI.00836-21. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

