Predominance of Dengue Virus Serotype-1/Genotype-I in Eastern and Southeastern Ethiopia
- PMID: 39205308
- PMCID: PMC11359325
- DOI: 10.3390/v16081334
Predominance of Dengue Virus Serotype-1/Genotype-I in Eastern and Southeastern Ethiopia
Abstract
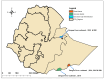
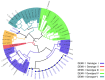
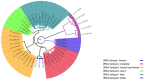
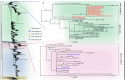
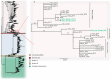
We determined the dengue virus (DENV) serotypes and genotypes in archived serum samples that were collected during the 2014-2016 and 2021 dengue outbreaks in Dire Dawa City and the Somali region in Ethiopia. DENV serotype 1 (DENV-1) was predominant followed by DENV serotype 2 (DENV-2). Thirteen of the DENV-1 strains were assigned to Genotype-I, while the remaining two were found to be Genotype-III. All three DENV-2 strains were assigned the Cosmopolitan Genotype. The DENV strains responsible for the outbreaks are genetically closely related to the DENV strains that circulated in neighboring and Asian countries. The findings also showed continued local transmission of a monophyletic lineage and a co-circulation of DENV-1 and DENV-2 during the outbreaks. There is a need to strengthen DENV genomic surveillance capacity for the early detection of circulating serotypes, and prevent devastating consequences of future outbreaks due to the co-circulation of different serotypes.
Keywords: Ethiopia; capsid/pre-membrane (CprM); dengue virus; genotype; outbreak; phylogenetic analysis; sequence; serotype.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Lineage B Genotype III of Dengue Virus Serotype 3 (DENV-3III_B) Is Responsible for Dengue Outbreak in Dire Dawa City, Ethiopia, 2023.Viruses. 2025 Feb 28;17(3):346. doi: 10.3390/v17030346. Viruses. 2025. PMID: 40143274 Free PMC article.
-
Circulating serotypes and genotypes of dengue virus during the 2023 outbreak in Eastern Nepal.J Clin Virol. 2024 Oct;174:105721. doi: 10.1016/j.jcv.2024.105721. Epub 2024 Aug 30. J Clin Virol. 2024. PMID: 39232301
-
Serotype and genomic diversity of dengue virus during the 2023 outbreak in Pakistan reveals the circulation of genotype III of DENV-1 and cosmopolitan genotype of DENV-2.J Med Virol. 2024 Jun;96(6):e29727. doi: 10.1002/jmv.29727. J Med Virol. 2024. PMID: 38864343
-
Genomic Characterization of Circulating Dengue Virus, Ethiopia, 2022-2023.Emerg Infect Dis. 2025 Mar;31(3):516-525. doi: 10.3201/eid3103.240996. Emerg Infect Dis. 2025. PMID: 40023801 Free PMC article.
-
Serotypic and genotypic profile of dengue virus outbreaks in India between 2011 and 2017.J Vector Borne Dis. 2025 Apr 1;62(2):135-142. doi: 10.4103/JVBD.JVBD_101_24. Epub 2024 Oct 5. J Vector Borne Dis. 2025. PMID: 39374060 Review.
Cited by
-
Lineage B Genotype III of Dengue Virus Serotype 3 (DENV-3III_B) Is Responsible for Dengue Outbreak in Dire Dawa City, Ethiopia, 2023.Viruses. 2025 Feb 28;17(3):346. doi: 10.3390/v17030346. Viruses. 2025. PMID: 40143274 Free PMC article.
-
Molecular Epidemiology of Travel-Associated and Locally Acquired Dengue Virus Infections in Catalonia, Spain, 2019.Viruses. 2025 Apr 26;17(5):621. doi: 10.3390/v17050621. Viruses. 2025. PMID: 40431634 Free PMC article.
References
-
- World Health Organization: WHO Dengue and Severe Dengue. Fact Sheet. March 2023. [(accessed on 11 August 2023)]. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- Woyessa A.B., Mengesha M., Kassa W., Kifle E., Wondabeku M., Girmay A., Kebede A., Jima D. The first acute febrile illness investigation associated with dengue fever in Ethiopia, 2013: A descriptive analysis. Ethiop. J. Health Dev. 2014;28:155–161.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

