Proteogenomic characterization of pancreatic neuroendocrine tumors uncovers hypoxia and immune signatures in clinically aggressive subtypes
- PMID: 39206147
- PMCID: PMC11350455
- DOI: 10.1016/j.isci.2024.110544
Proteogenomic characterization of pancreatic neuroendocrine tumors uncovers hypoxia and immune signatures in clinically aggressive subtypes
Abstract
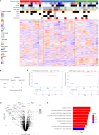
Pancreatic neuroendocrine tumors (PanNETs) represent well-differentiated endocrine neoplasms with variable clinical outcomes. Predicting patient outcomes using the current tumor grading system is challenging. In addition, traditional systemic treatment options for PanNETs, such as somatostatin analogs or cytotoxic chemotherapies, are very limited. To address these issues, we characterized PanNETs using integrated proteogenomics and identified four subtypes. Two proteomic subtypes showed high recurrence rates, suggesting clinical aggressiveness that was missed by current classification. Hypoxia and inflammatory pathways were significantly enriched in the clinically aggressive subtypes. Detailed analyses revealed metabolic adaptation via glycolysis upregulation and oxidative phosphorylation downregulation under hypoxic conditions. Inflammatory signature analysis revealed that immunosuppressive molecules were enriched in immune hot tumors and might be immunotherapy targets. In this study, we characterized clinically aggressive proteomic subtypes of well-differentiated PanNETs and identified candidate therapeutic targets.
Keywords: Cancer; Cancer systems biology; Genomics; Proteomics.
© 2024 Published by Elsevier Inc.
Conflict of interest statement
D.S.K. is now an employee of Paige.AI. M.H.R. is a member of the Scientific Advisory Boards of Azenta Life Sciences and Universal Diagnostics (UDX). None of these companies had any role in the support, design, execution, data analysis, or any other aspect of this study.
Figures





References
-
- Yao J.C., Hassan M., Phan A., Dagohoy C., Leary C., Mares J.E., Abdalla E.K., Fleming J.B., Vauthey J.N., Rashid A., Evans D.B. One hundred years after "carcinoid": epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J. Clin. Oncol. 2008;26:3063–3072. doi: 10.1200/jco.2007.15.4377. - DOI - PubMed
-
- Digestive System Tumours . 5th Edition. 2019. WHO Classification of Tumours.
-
- Yang M., Zeng L., Ke N.W., Tan C.L., Tian B.L., Liu X.B., Xiang B., Zhang Y. World Health Organization grading classification for pancreatic neuroendocrine neoplasms: a comprehensive analysis from a large Chinese institution. BMC Cancer. 2020;20:906. doi: 10.1186/s12885-020-07356-5. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

