Transient Inhibition of Translation Improves Cardiac Function After Ischemia/Reperfusion by Attenuating the Inflammatory Response
- PMID: 39206545
- PMCID: PMC11472906
- DOI: 10.1161/CIRCULATIONAHA.123.067479
Transient Inhibition of Translation Improves Cardiac Function After Ischemia/Reperfusion by Attenuating the Inflammatory Response
Abstract
Background: The myocardium adapts to ischemia/reperfusion (I/R) by changes in gene expression, determining the cardiac response to reperfusion. mRNA translation is a key component of gene expression. It is largely unknown how regulation of mRNA translation contributes to cardiac gene expression and inflammation in response to reperfusion and whether it can be targeted to mitigate I/R injury.
Methods: To examine translation and its impact on gene expression in response to I/R, we measured protein synthesis after reperfusion in vitro and in vivo. Underlying mechanisms of translational control were examined by pharmacological and genetic targeting of translation initiation in mice. Cell type-specific ribosome profiling was performed in mice that had been subjected to I/R to determine the impact of mRNA translation on the regulation of gene expression in cardiomyocytes. Translational regulation of inflammation was studied by quantification of immune cell infiltration, inflammatory gene expression, and cardiac function after short-term inhibition of translation initiation.
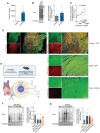
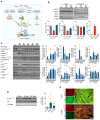
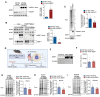
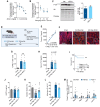
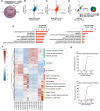
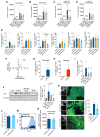
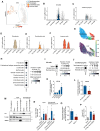
Results: Reperfusion induced a rapid recovery of translational activity that exceeds baseline levels in the infarct and border zone and is mediated by translation initiation through the mTORC1 (mechanistic target of rapamycin complex 1)-4EBP1 (eIF4E-binding protein 1)-eIF (eukaryotic initiation factor) 4F axis. Cardiomyocyte-specific ribosome profiling identified that I/R increased translation of mRNA networks associated with cardiac inflammation and cell infiltration. Short-term inhibition of the mTORC1-4EBP1-eIF4F axis decreased the expression of proinflammatory cytokines such as Ccl2 (C-C motif chemokine ligand 2) of border zone cardiomyocytes, thereby attenuating Ly6Chi monocyte infiltration and myocardial inflammation. In addition, we identified a systemic immunosuppressive effect of eIF4F translation inhibitors on circulating monocytes, directly inhibiting monocyte infiltration. Short-term pharmacological inhibition of eIF4F complex formation by 4EGI-1 or rapamycin attenuated translation, reduced infarct size, and improved cardiac function after myocardial infarction.
Conclusions: Global protein synthesis is inhibited during ischemia and shortly after reperfusion, followed by a recovery of protein synthesis that exceeds baseline levels in the border and infarct zones. Activation of mRNA translation after reperfusion is driven by mTORC1/eIF4F-mediated regulation of initiation and mediates an mRNA network that controls inflammation and monocyte infiltration to the myocardium. Transient inhibition of the mTORC1-/eIF4F axis inhibits translation and attenuates Ly6Chi monocyte infiltration by inhibiting a proinflammatory response at the site of injury and of circulating monocytes.
Keywords: TOR serine-threonine kinases; chemokine CCL2; eukaryotic initiation factor-4F; inflammation; myocardial infarction; peptide chain initiation, translational; reperfusion injury.
Conflict of interest statement
None.
Figures

References
-
- Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS, Boehme AK, Buxton AE, Carson AP, Commodore-Mensah Y, et al. Heart disease and stroke statistics—2022 update: a report from the American Heart Association. Circulation. 2022;145:e153–e639. doi: 10.1161/CIR.0000000000001052 - PubMed
-
- Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, Caforio ALP, Crea F, Goudevenos JA, Halvorsen S, et al. ; ESC Scientific Document Group. 2017 ESC guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: the Task Force for the Management of Acute Myocardial Infarction in Patients Presenting with ST-Segment Elevation of the European Society of Cardiology (ESC). Eur Heart J. 2018;39:119–177. doi: 10.1093/eurheartj/ehx393 - PubMed
-
- Heusch G. Myocardial ischaemia–reperfusion injury and cardioprotection in perspective. Nat Rev Cardiol. 2020;17:773–789. doi: 10.1038/s41569-020-0403-y - PubMed
-
- Algoet M, Janssens S, Himmelreich U, Gsell W, Pusovnik M, Van den Eynde J, Oosterlinck W. Myocardial ischemia-reperfusion injury and the influence of inflammation. Trends Cardiovasc Med. 2023;33:357–366. doi: 10.1016/j.tcm.2022.02.005 - PubMed
-
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

