Evolutionary history and origins of Dsr-mediated sulfur oxidation
- PMID: 39206688
- PMCID: PMC11406059
- DOI: 10.1093/ismejo/wrae167
Evolutionary history and origins of Dsr-mediated sulfur oxidation
Abstract
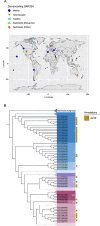
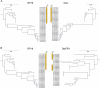
Microorganisms play vital roles in sulfur cycling through the oxidation of elemental sulfur and reduction of sulfite. These metabolisms are catalyzed by dissimilatory sulfite reductases (Dsr) functioning in either the reductive or reverse, oxidative direction. Dsr-mediated sulfite reduction is an ancient metabolism proposed to have fueled energy metabolism in some of Earth's earliest microorganisms, whereas sulfur oxidation is believed to have evolved later in association with the widespread availability of oxygen on Earth. Organisms are generally believed to carry out either the reductive or oxidative pathway, yet organisms from diverse phyla have been discovered with gene combinations that implicate them in both pathways. A comprehensive investigation into the metabolisms of these phyla regarding Dsr is currently lacking. Here, we selected one of these phyla, the metabolically versatile candidate phylum SAR324, to study the ecology and evolution of Dsr-mediated metabolism. We confirmed that diverse SAR324 encode genes associated with reductive Dsr, oxidative Dsr, or both. Comparative analyses with other Dsr-encoding bacterial and archaeal phyla revealed that organisms encoding both reductive and oxidative Dsr proteins are constrained to a few phyla. Further, DsrAB sequences from genomes belonging to these phyla are phylogenetically positioned at the interface between well-defined oxidative and reductive bacterial clades. The phylogenetic context and dsr gene content in these organisms points to an evolutionary transition event that ultimately gave way to oxidative Dsr-mediated metabolism. Together, this research suggests that SAR324 and other phyla with mixed dsr gene content are associated with the evolution and origins of Dsr-mediated sulfur oxidation.
Keywords: SAR324; dissimilatory sulfite reductase; metagenomics; sulfur oxidation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Georgoulias AK, Balis D, Koukouli MEet al. A study of the total atmospheric sulfur dioxide load using ground-based measurements and the satellite derived sulfur dioxide index. Atmos Environ 2009;43:1693–701.
-
- Ksionzek KB, Lechtenfeld OJ, McCallister SLet al. Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory. Science 2016;354:456–9. - PubMed
-
- Schoenau JJ, Malhi SS. Sulfur forms and cycling processes in soil and their relationship to sulfur fertility. In: Jez J. (ed.), Agronomy Monographs. Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, 2015, 1–10.
-
- Wedepohl KH. Sulfur in the Earth’s crust, its origin and natural cycle. In: Müller A, Krebs B. (eds.), Studies in Inorganic Chemistry. Elsevier, 1984, 39–54.
-
- Brosnan JT, Brosnan ME. The sulfur-containing amino acids: an overview. J Nutr 2006;136:1636S–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

