Hypoxic Upregulation of IER2 Increases Paracrine GMFG Signaling of Endoplasmic Reticulum Stress-CAF to Promote Chordoma Progression via Targeting ITGB1
- PMID: 39207055
- PMCID: PMC11515918
- DOI: 10.1002/advs.202405421
Hypoxic Upregulation of IER2 Increases Paracrine GMFG Signaling of Endoplasmic Reticulum Stress-CAF to Promote Chordoma Progression via Targeting ITGB1
Abstract
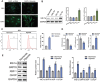
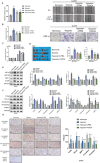
Currently, the oncogenic mechanism of endoplasmic reticulum stress-CAF (ERS-CAF) subpopulation in chordoma remains unknown. Here, single-cell RNA sequencing, spatial transcriptomics, GeoMx Digital Spatial Profiler, data-independent acquisition proteomics, bulk RNA-seq, and multiplexed quantitative immunofluorescence are used to unveil the precise molecular mechanism of how ERS-CAF affected chordoma progression. Results show that hypoxic microenvironment reprograms CAFs into ERS-CAF subtype. Mechanistically, this occurrs via hypoxia-mediated transcriptional upregulation of IER2. Overexpression of IER2 in CAFs promotes chordoma progression, which can be impeded by IER2 knockdown or use of ERS inhibitors. IER2 also induces expression of ERS-CAF marker genes and results in production of a pro-tumorigenic paracrine GMFG signaling, which exert its biological function via directly binding to ITGB1 on tumor cells. ITGB1 inhibition attenuates tumor malignant progression, which can be partially reversed by exogenous GMFG intervention. Further analyses reveal a positive correlation between ITGB1high tumor cell counts and SPP1+ macrophage density, as well as the spatial proximity of these two cell types. Clinically, a significant correlation of high IER2/ITGB1 expression with tumor aggressive phenotype and poor patient survival is observed. Collectively, the findings suggest that ERS-CAF regulates SPP1+ macrophage to aggravate chordoma progression via the IER2/GMFG/ITGB1 axis, which may be targeted therapeutically in future.
Keywords: IER2/GMFG/ITGB1 axis; cancer‐associated fibroblasts; chordoma progression; endoplasmic reticulum stress; hypoxia; paracrine.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
- 82003802/National Natural Science Foundation of China
- 82002364/National Natural Science Foundation of China
- 202106370071/China Scholarship Council
- 2019JJ50542/Natural Science Foundation of Hunan Province
- 2023JJ50156/Natural Science Foundation of Hunan Province
- 2023JJ40596/Natural Science Foundation of Hunan Province
- 2023JJ40587/Natural Science Foundation of Hunan Province
- 2021JJ40509/Natural Science Foundation of Hunan Province
- 2023JJ20035/Hunan Provincial Natural Science Foundation for Excellent Youth Scholars
- 2023RC3172/Science and Technology Innovation Program of Hunan Province
- 20224310NHYCG04/Clinical Research 4310 Program of the First Affiliated Hospital of the University of South China
- 20224310NHYCG03/Clinical Research 4310 Program of the First Affiliated Hospital of the University of South China
- 20201978/Project for Clinical Research of Hunan Provincial Health Commission
- 20201962/Project for Clinical Research of Hunan Provincial Health Commission
- 20201956/Project for Clinical Research of Hunan Provincial Health Commission
- 22B0441/Research Foundation of Education Bureau of Hunan Province
- Xiaohe Technology Talent Project of Hengyang City
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
