Mutant induced neurons and humanized mice enable identification of Niemann-Pick type C1 proteostatic therapies
- PMID: 39207850
- PMCID: PMC11530122
- DOI: 10.1172/jci.insight.179525
Mutant induced neurons and humanized mice enable identification of Niemann-Pick type C1 proteostatic therapies
Abstract
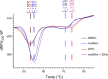
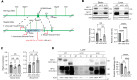
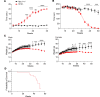
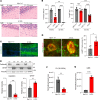
Therapeutics that rescue folding, trafficking, and function of disease-causing missense mutants are sought for a host of human diseases, but efforts to leverage model systems to test emerging strategies have met with limited success. Such is the case for Niemann-Pick type C1 disease, a lysosomal disorder characterized by impaired intracellular cholesterol trafficking, progressive neurodegeneration, and early death. NPC1, a multipass transmembrane glycoprotein, is synthesized in the endoplasmic reticulum and traffics to late endosomes/lysosomes, but this process is often disrupted in disease. We sought to identify small molecules that promote folding and enable lysosomal localization and functional recovery of mutant NPC1. We leveraged a panel of isogenic human induced neurons expressing distinct NPC1 missense mutations. We used this panel to rescreen compounds that were reported previously to correct NPC1 folding and trafficking. We established mo56-hydroxycholesterol (mo56Hc) as a potent pharmacological chaperone for several NPC1 mutants. Furthermore, we generated mice expressing human I1061T NPC1, a common mutation in patients. We demonstrated that this model exhibited disease phenotypes and recapitulated the protein trafficking defects, lipid storage, and response to mo56Hc exhibited by human cells expressing I1061T NPC1. These tools established a paradigm for testing and validation of proteostatic therapeutics as an important step toward the development of disease-modifying therapies.
Keywords: Lysosomes; Neurological disorders; Neuroscience; Protein misfolding.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

