Decoding the Foodome: Molecular Networks Connecting Diet and Health
- PMID: 39207880
- PMCID: PMC11610447
- DOI: 10.1146/annurev-nutr-062322-030557
Decoding the Foodome: Molecular Networks Connecting Diet and Health
Abstract
Diet, a modifiable risk factor, plays a pivotal role in most diseases, from cardiovascular disease to type 2 diabetes mellitus, cancer, and obesity. However, our understanding of the mechanistic role of the chemical compounds found in food remains incomplete. In this review, we explore the "dark matter" of nutrition, going beyond the macro- and micronutrients documented by national databases to unveil the exceptional chemical diversity of food composition. We also discuss the need to explore the impact of each compound in the presence of associated chemicals and relevant food sources and describe the tools that will allow us to do so. Finally, we discuss the role of network medicine in understanding the mechanism of action of each food molecule. Overall, we illustrate the important role of network science and artificial intelligence in our ability to reveal nutrition's multifaceted role in health and disease.
Keywords: artificial intelligence; complexity; machine learning; network medicine; network science; nutrition; systems pharmacology.
Figures




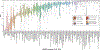
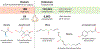

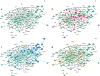


References
-
- Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, et al. 2012. KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant Cell Physiol. 53(2):e1. - PubMed
-
- Aguilera JM. 2019. The food matrix: implications in processing, nutrition and health. Crit. Rev. Food Sci. Nutr 59(22):3612–29 - PubMed
-
- Alonso-Pedrero L, Ojeda-Rodríguez A, Martínez-González MA, Zalba G, Bes-Rastrollo M, Marti A. 2020. Ultra-processed food consumption and the risk of short telomeres in an elderly population of the Seguimiento Universidad de Navarra (SUN) Project. Am. J. Clin. Nutr 111(6):1259–66 - PubMed
-
- Int AOAC. 2023. Official Methods of Analysis of AOAC International, Vols. 1–2. Washington, DC: Oxford Univ. Press. 22nd ed.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

