Marine sponge microbe provides insights into evolution and virulence of the tubercle bacillus
- PMID: 39207937
- PMCID: PMC11361433
- DOI: 10.1371/journal.ppat.1012440
Marine sponge microbe provides insights into evolution and virulence of the tubercle bacillus
Abstract
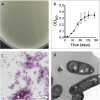
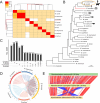
Reconstructing the evolutionary origins of Mycobacterium tuberculosis, the causative agent of human tuberculosis, has helped identify bacterial factors that have led to the tubercle bacillus becoming such a formidable human pathogen. Here we report the discovery and detailed characterization of an exceedingly slow growing mycobacterium that is closely related to M. tuberculosis for which we have proposed the species name Mycobacterium spongiae sp. nov., (strain ID: FSD4b-SM). The bacterium was isolated from a marine sponge, taken from the waters of the Great Barrier Reef in Queensland, Australia. Comparative genomics revealed that, after the opportunistic human pathogen Mycobacterium decipiens, M. spongiae is the most closely related species to the M. tuberculosis complex reported to date, with 80% shared average nucleotide identity and extensive conservation of key M. tuberculosis virulence factors, including intact ESX secretion systems and associated effectors. Proteomic and lipidomic analyses showed that these conserved systems are functional in FSD4b-SM, but that it also produces cell wall lipids not previously reported in mycobacteria. We investigated the virulence potential of FSD4b-SM in mice and found that, while the bacteria persist in lungs for 56 days after intranasal infection, no overt pathology was detected. The similarities with M. tuberculosis, together with its lack of virulence, motivated us to investigate the potential of FSD4b-SM as a vaccine strain and as a genetic donor of the ESX-1 genetic locus to improve BCG immunogenicity. However, neither of these approaches resulted in superior protection against M. tuberculosis challenge compared to BCG vaccination alone. The discovery of M. spongiae adds to our understanding of the emergence of the M. tuberculosis complex and it will be another useful resource to refine our understanding of the factors that shaped the evolution and pathogenesis of M. tuberculosis.
Copyright: © 2024 Pidot et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- WHO. Global Tuberculosis Report 2022. Geneva: World Health Organisation; 2022.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

