A p62-dependent rheostat dictates micronuclei catastrophe and chromosome rearrangements
- PMID: 39208097
- PMCID: PMC11664475
- DOI: 10.1126/science.adj7446
A p62-dependent rheostat dictates micronuclei catastrophe and chromosome rearrangements
Abstract
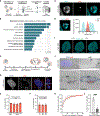
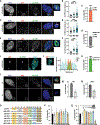
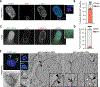
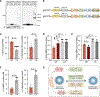
Chromosomal instability (CIN) generates micronuclei-aberrant extranuclear structures that catalyze the acquisition of complex chromosomal rearrangements present in cancer. Micronuclei are characterized by persistent DNA damage and catastrophic nuclear envelope collapse, which exposes DNA to the cytoplasm. We found that the autophagic receptor p62/SQSTM1 modulates micronuclear stability, influencing chromosome fragmentation and rearrangements. Mechanistically, proximity of micronuclei to mitochondria led to oxidation-driven homo-oligomerization of p62, limiting endosomal sorting complex required for transport (ESCRT)-dependent micronuclear envelope repair by triggering autophagic degradation. We also found that p62 levels correlate with increased chromothripsis across human cancer cell lines and with increased CIN in colorectal tumors. Thus, p62 acts as a regulator of micronuclei and may serve as a prognostic marker for tumors with high CIN.
Conflict of interest statement
Figures




Comment in
-
Micronuclear collapse mechanisms in cancer.Science. 2024 Aug 30;385(6712):930-931. doi: 10.1126/science.adr7417. Epub 2024 Aug 29. Science. 2024. PMID: 39208121
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

