Aggregate-selective removal of pathological tau by clustering-activated degraders
- PMID: 39208111
- PMCID: PMC7616837
- DOI: 10.1126/science.adp5186
Aggregate-selective removal of pathological tau by clustering-activated degraders
Abstract
Selective degradation of pathological protein aggregates while sparing monomeric forms is of major therapeutic interest. The E3 ligase tripartite motif-containing protein 21 (TRIM21) degrades antibody-bound proteins in an assembly state-specific manner due to the requirement of TRIM21 RING domain clustering for activation, yet effective targeting of intracellular assemblies remains challenging. Here, we fused the RING domain of TRIM21 to a target-specific nanobody to create intracellularly expressed constructs capable of selectively degrading assembled proteins. We evaluated this approach against green fluorescent protein-tagged histone 2B (H2B-GFP) and tau, a protein that undergoes pathological aggregation in Alzheimer's and other neurodegenerative diseases. RING-nanobody degraders prevented or reversed tau aggregation in culture and in vivo, with minimal impact on monomeric tau. This approach may have therapeutic potential for the many disorders driven by intracellular protein aggregation.
Conflict of interest statement
W.A.M and L.C.J are academic founders, shareholders and scientific advisors for TRIMTECH Therapeutics.
Figures

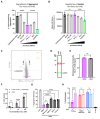
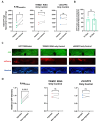
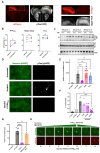
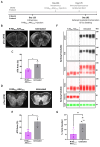
References
-
- Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, Castellani RJ, Crain BJ, Davies P, Del Tredici K, Duyckaerts C, et al. Correlation of Alzheimer Disease Neuropathologic Changes With Cognitive Status: A Review of the Literature. J Neuropathol Exp Neurol. 2012;71:362–381. doi: 10.1097/NEN.0b013e31825018f7. - DOI - PMC - PubMed
-
- Hanseeuw B, Jacobs HIL, Becker A, Buckley RF, Properzi MJ, Farrell ME, Schultz AP, Sanchez JS, Chhatwal JP, Price JC, Sperling RA, et al. Longitudinal associations between amyloid and tau-PET: Impact for prevention trials. Alzheimer’s & Dementia. 2021;17
-
- DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, Wegener AJ, Chen G, Shen T, Tran H, Nichols B, Zanardi TA, et al. Tau Reduction Prevents Neuronal Loss and Reverses Pathological Tau Deposition and Seeding in Mice with Tauopathy. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aag0481. - DOI - PMC - PubMed
-
- Edwards AL, Collins JA, Junge C, Kordasiewicz H, Mignon L, Wu S, Li Y, Lin L, DuBois J, Hutchison RM, Ziogas N, et al. Exploratory Tau Biomarker Results From a Multiple Ascending-Dose Study of BIIB080 in Alzheimer Disease. JAMA Neurol. 2023;80:1344. doi: 10.1001/jamaneurol.2023.3861. - DOI - PMC - PubMed
-
- Mummery CJ, Børjesson-Hanson A, Blackburn DJ, Vijverberg EGB, De Deyn PP, Ducharme S, Jonsson M, Schneider A, Rinne JO, Ludolph AC, Bodenschatz R, et al. Tau-targeting antisense oligonucleotide MAPTRx in mild Alzheimer’s disease: a phase 1b, randomized, placebo-controlled trial. Nature Medicine. 2023;29:1437–1447. doi: 10.1038/s41591-023-02326-3. 2023 29:6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

