Apobec-mediated retroviral hypermutation in vivo is dependent on mouse strain
- PMID: 39208378
- PMCID: PMC11389910
- DOI: 10.1371/journal.ppat.1012505
Apobec-mediated retroviral hypermutation in vivo is dependent on mouse strain
Abstract
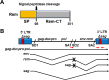
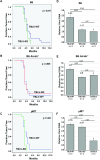
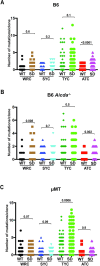
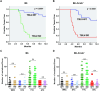
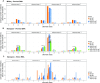
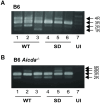
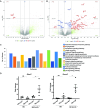
Replication of the complex retrovirus mouse mammary tumor virus (MMTV) is antagonized by murine Apobec3 (mA3), a member of the Apobec family of cytidine deaminases. We have shown that MMTV-encoded Rem protein inhibits proviral mutagenesis by the Apobec enzyme, activation-induced cytidine deaminase (AID) during viral replication in BALB/c mice. To further study the role of Rem in vivo, we have infected C57BL/6 (B6) mice with a superantigen-independent lymphomagenic strain of MMTV (TBLV-WT) or a mutant strain that is defective in Rem and its cleavage product Rem-CT (TBLV-SD). Compared to BALB/c, B6 mice were more susceptible to TBLV infection and tumorigenesis. Furthermore, unlike MMTV, TBLV induced T-cell tumors in B6 μMT mice, which lack membrane-bound IgM and conventional B-2 cells. At limiting viral doses, loss of Rem expression in TBLV-SD-infected B6 mice accelerated tumorigenesis compared to TBLV-WT in either wild-type B6 or AID-knockout mice. Unlike BALB/c results, high-throughput sequencing indicated that proviral G-to-A or C-to-T mutations were unchanged regardless of Rem expression in B6 tumors. However, knockout of both AID and mA3 reduced G-to-A mutations. Ex vivo stimulation showed higher levels of mA3 relative to AID in B6 compared to BALB/c splenocytes, and effects of agonists differed in the two strains. RNA-Seq revealed increased transcripts related to growth factor and cytokine signaling in TBLV-SD-induced tumors relative to TBLV-WT-induced tumors, consistent with another Rem function. Thus, Rem-mediated effects on tumorigenesis in B6 mice are independent of Apobec-mediated proviral hypermutation.
Copyright: © 2024 Byun et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Update of
-
Apobec-Mediated Retroviral Hypermutation In Vivo is Dependent on Mouse Strain.bioRxiv [Preprint]. 2023 Nov 2:2023.11.02.565355. doi: 10.1101/2023.11.02.565355. bioRxiv. 2023. Update in: PLoS Pathog. 2024 Aug 29;20(8):e1012505. doi: 10.1371/journal.ppat.1012505. PMID: 37961113 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

