Proteo-metabolomics and patient tumor slice experiments point to amino acid centrality for rewired mitochondria in fibrolamellar carcinoma
- PMID: 39208801
- PMCID: PMC11528240
- DOI: 10.1016/j.xcrm.2024.101699
Proteo-metabolomics and patient tumor slice experiments point to amino acid centrality for rewired mitochondria in fibrolamellar carcinoma
Abstract
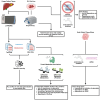
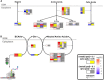
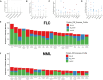
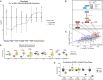
Fibrolamellar carcinoma (FLC) is a rare, lethal, early-onset liver cancer with a critical need for new therapeutics. The primary driver in FLC is the fusion oncoprotein, DNAJ-PKAc, which remains challenging to target therapeutically. It is critical, therefore, to expand understanding of the FLC molecular landscape to identify druggable pathways/targets. Here, we perform the most comprehensive integrative proteo-metabolomic analysis of FLC. We also conduct nutrient manipulation, respirometry analyses, as well as key loss-of-function assays in FLC tumor tissue slices from patients. We propose a model of cellular energetics in FLC pointing to proline anabolism being mediated by ornithine aminotransferase hyperactivity and ornithine transcarbamylase hypoactivity with serine and glutamine catabolism fueling the process. We highlight FLC's potential dependency on voltage-dependent anion channel (VDAC), a mitochondrial gatekeeper for anions including pyruvate. The metabolic rewiring in FLC that we propose in our model, with an emphasis on mitochondria, can be exploited for therapeutic vulnerabilities.
Keywords: alpha-ketoglutarate; fibrolamellar carcinoma; glucose; glutamine; metabolomics; mitochondria; proline; proteomics; pyruvate; serine.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Dinh T.A., Utria A.F., Barry K.C., Ma R., Abou-Alfa G.K., Gordan J.D., Jaffee E.M., ’Sco’t J.D., Zucman-Rossi J., O'Neill A.F., et al. A framework for fibrolamellar carcinoma research and clinical trials. Nat. Rev. Gastroenterol. Hepatol. 2022;19:328–342. doi: 10.1038/s41575-022-00580-3. - DOI - PMC - PubMed

