Genome access is transcription factor-specific and defined by nucleosome position
- PMID: 39208807
- PMCID: PMC11420395
- DOI: 10.1016/j.molcel.2024.08.009
Genome access is transcription factor-specific and defined by nucleosome position
Abstract
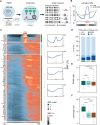
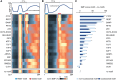
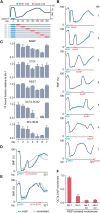
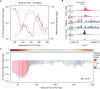
Mammalian gene expression is controlled by transcription factors (TFs) that engage sequence motifs in a chromatinized genome, where nucleosomes can restrict DNA access. Yet, how nucleosomes affect individual TFs remains unclear. Here, we measure the ability of over one hundred TF motifs to recruit TFs in a defined chromosomal locus in mouse embryonic stem cells. This identifies a set sufficient to enable the binding of TFs with diverse tissue specificities, functions, and DNA-binding domains. These chromatin-competent factors are further classified when challenged to engage motifs within a highly phased nucleosome. The pluripotency factors OCT4-SOX2 preferentially engage non-nucleosomal and entry-exit motifs, but not nucleosome-internal sites, a preference that also guides binding genome wide. By contrast, factors such as BANP, REST, or CTCF engage throughout, causing nucleosomal displacement. This supports that TFs vary widely in their sensitivity to nucleosomes and that genome access is TF specific and influenced by nucleosome position in the cell.
Keywords: accessibility; chromatin; genome; nucleosome; pioneer factors; transcription factors.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

