BSAlign: A Library for Nucleotide Sequence Alignment
- PMID: 39209796
- PMCID: PMC12016559
- DOI: 10.1093/gpbjnl/qzae025
BSAlign: A Library for Nucleotide Sequence Alignment
Abstract
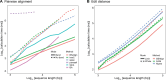
Increasing the accuracy of the nucleotide sequence alignment is an essential issue in genomics research. Although classic dynamic programming (DP) algorithms (e.g., Smith-Waterman and Needleman-Wunsch) guarantee to produce the optimal result, their time complexity hinders the application of large-scale sequence alignment. Many optimization efforts that aim to accelerate the alignment process generally come from three perspectives: redesigning data structures [e.g., diagonal or striped Single Instruction Multiple Data (SIMD) implementations], increasing the number of parallelisms in SIMD operations (e.g., difference recurrence relation), or reducing search space (e.g., banded DP). However, no methods combine all these three aspects to build an ultra-fast algorithm. In this study, we developed a Banded Striped Aligner (BSAlign) library that delivers accurate alignment results at an ultra-fast speed by knitting a series of novel methods together to take advantage of all of the aforementioned three perspectives with highlights such as active F-loop in striped vectorization and striped move in banded DP. We applied our new acceleration design on both regular and edit distance pairwise alignment. BSAlign achieved 2-fold speed-up than other SIMD-based implementations for regular pairwise alignment, and 1.5-fold to 4-fold speed-up in edit distance-based implementations for long reads. BSAlign is implemented in C programing language and is available at https://github.com/ruanjue/bsalign.
Keywords: Banded dynamic programming; Edit distance; F evaluation; Pairwise alignment; Striped vectorization.
© The Author(s) 2024. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Conflict of interest statement
Both authors have declared no competing interests.
Figures


References
-
- Needleman SB, Wunsch CD.. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 1970;48:443–53. - PubMed
-
- Smith T, Waterman M.. Identification of common molecular subsequences. J Mol Biol 1981;147:195–7. - PubMed
-
- Wozniak A. Using video-oriented instructions to speed up sequence comparison. Comput Appl Biosci 1997;13:145–50. - PubMed
-
- Rognes T, Seeberg E.. Six-fold speed-up of Smith–Waterman sequence database searches using parallel processing on common microprocessors. Bioinformatics 2000;16:699–706. - PubMed
-
- Zhang J, Lan H, Chan Y, Shang Y, Schmidt B, Liu W.. BGSA: a bit-parallel global sequence alignment toolkit for multi-core and many-core architectures. Bioinformatics 2019;35:2306–8. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

