Helical ultrastructure of the L-ENA spore aggregation factor of a Bacillus paranthracis foodborne outbreak strain
- PMID: 39209852
- PMCID: PMC11362473
- DOI: 10.1038/s41467-024-51804-w
Helical ultrastructure of the L-ENA spore aggregation factor of a Bacillus paranthracis foodborne outbreak strain
Abstract
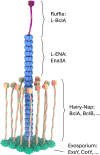
In pathogenic Bacillota, spores can form an infectious particle and can take up a central role in the environmental persistence and dissemination of disease. A poorly understood aspect of spore-mediated infection is the fibrous structures or 'endospore appendages' (ENAs) that have been seen to decorate the spores of pathogenic Bacilli and Clostridia. Current methodological approaches are opening a window on these long enigmatic structures. Using cryoID, Alphafold modelling and genetic approaches we identify a sub-class of robust ENAs in a Bacillus paranthracis foodborne outbreak strain. We demonstrate that L-ENA are encoded by a rare three-gene cluster (ena3) that contains all components for the self-assembly of ladder-like protein nanofibers of stacked heptameric rings, their anchoring to the exosporium, and their termination in a trimeric 'ruffle' made of a complement C1Q-like BclA paralogue. The role of ENA fibers in spore-spore interaction and the distribution of L-ENA operon as mobile genetic elements in B. cereus s.l. strains suggest that L-ENA fibers may increase the survival, spread and virulence of these strains.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

