Spaceflight alters host-gut microbiota interactions
- PMID: 39209868
- PMCID: PMC11362537
- DOI: 10.1038/s41522-024-00545-1
Spaceflight alters host-gut microbiota interactions
Abstract
The ISS rodent habitat has provided crucial insights into the impact of spaceflight on mammals, inducing symptoms characteristic of liver disease, insulin resistance, osteopenia, and myopathy. Although these physiological responses can involve the microbiome on Earth, host-microbiota interactions during spaceflight are still being elucidated. We explore murine gut microbiota and host gene expression in the colon and liver after 29 and 56 days of spaceflight using multiomics. Metagenomics revealed significant changes in 44 microbiome species, including relative reductions in bile acid and butyrate metabolising bacteria like Extibacter muris and Dysosmobacter welbionis. Functional prediction indicate over-representation of fatty acid and bile acid metabolism, extracellular matrix interactions, and antibiotic resistance genes. Host gene expression described corresponding changes to bile acid and energy metabolism, and immune suppression. These changes imply that interactions at the host-gut microbiome interface contribute to spaceflight pathology and that these interactions might critically influence human health and long-duration spaceflight feasibility.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
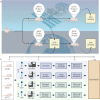
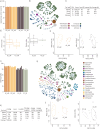



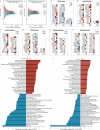
References
-
- ISECG. 3. The International Space Exploration Coordination Group.https://www.globalspaceexploration.org/ (2018).
-
- ISECG. Washington National Aeronautics and Space Administration Headquarters.https://www.nasa.gov (2022).
-
- National Aeronautics and Space Administration (NASA). Fiscal Year 2023 Budget Estimates. Retrieved from https://www.nasa.gov/wp-content/uploads/2023/07/nasa-fy-2023-agencyfact-... (Washington, D.C.: NASA, 2022).
-
- National Aeronautics and Space Administration (NASA). Moon to Mars Objectives. Washington, D.C.:NASA. Retrieved from https://www.nasa.gov/sites/default/files/atoms/files/m2m-objectives-exec... (Washington, D.C.:NASA, 2022).
-
- Stein, T., Schulter, M. & Boden, G. Development of insulin resistance by astronauts during spaceflight. Aviat. Space Environ. Med.65, 1091–1096 (1994). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

