Effect of breed and dietary composition on the miRNA profile of beef steers divergent for feed efficiency
- PMID: 39209905
- PMCID: PMC11362461
- DOI: 10.1038/s41598-024-70669-z
Effect of breed and dietary composition on the miRNA profile of beef steers divergent for feed efficiency
Abstract
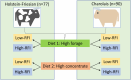
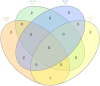
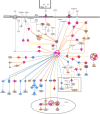
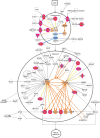
Identifying and breeding cattle that are more feed efficient is of great benefit to beef production. Additionally, it is crucial that genes contributing to feed efficiency are robust across varying management settings including dietary source as well as being relevant across contrasting breeds of cattle. The aim of this study was to determine miRNAs that are contributing to the expression of residual feed intake (RFI) across two breeds and dietary sources. miRNA profiling was undertaken in Longissimus dorsi tissue of Charolais and Holstein-Friesian steers divergent for RFI phenotype following two contrasting consecutive diets (high-forage and high-concentrate). Ten miRNA were identified as differentially expressed (adj. P < 0.1) across the breed and diet contrasts examined. Of particular interest was the differential expression of miR-2419-5p and miR-2415-3p, both of which were up-regulated in the Low-RFI Charolais steers across each dietary phase. Pathway analysis of target mRNA genes of differentially expressed miRNA revealed enrichment (P < 0.05) for pathways including metabolic related pathways, insulin receptor signalling, adipogenesis as well as pathways related to skeletal muscle growth. These results provide insight into the skeletal muscle miRNAome of beef cattle and their potential molecular regulatory mechanisms relating to feed efficiency in beef cattle.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Manafiazar, G. et al. Methane and carbon dioxide emissions from yearling beef heifers and mature cows classified for residual feed intake under drylot conditions. Can. J. Anim. Sci.100, 522–535 (2020). 10.1139/cjas-2019-0032 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

