Global 13C tracing and metabolic flux analysis of intact human liver tissue ex vivo
- PMID: 39210089
- PMCID: PMC11496108
- DOI: 10.1038/s42255-024-01119-3
Global 13C tracing and metabolic flux analysis of intact human liver tissue ex vivo
Abstract
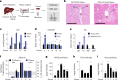
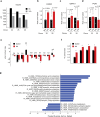
Liver metabolism is central to human physiology and influences the pathogenesis of common metabolic diseases. Yet, our understanding of human liver metabolism remains incomplete, with much of current knowledge based on animal or cell culture models that do not fully recapitulate human physiology. Here, we perform in-depth measurement of metabolism in intact human liver tissue ex vivo using global 13C tracing, non-targeted mass spectrometry and model-based metabolic flux analysis. Isotope tracing allowed qualitative assessment of a wide range of metabolic pathways within a single experiment, confirming well-known features of liver metabolism but also revealing unexpected metabolic activities such as de novo creatine synthesis and branched-chain amino acid transamination, where human liver appears to differ from rodent models. Glucose production ex vivo correlated with donor plasma glucose, suggesting that cultured liver tissue retains individual metabolic phenotypes, and could be suppressed by postprandial levels of nutrients and insulin, and also by pharmacological inhibition of glycogen utilization. Isotope tracing ex vivo allows measuring human liver metabolism with great depth and resolution in an experimentally tractable system.
© 2024. The Author(s).
Conflict of interest statement
M.J. is an employee at Sapient Bioanalytics, and holds equity in the company. The other authors declare no competing interests.
Figures








Similar articles
-
Simultaneous tracing of carbon and nitrogen isotopes in human cells.Mol Biosyst. 2016 May 24;12(6):1929-37. doi: 10.1039/c6mb00009f. Mol Biosyst. 2016. PMID: 27098229 Free PMC article.
-
Metabolic pathway analysis of tumors using stable isotopes.Semin Cancer Biol. 2025 Aug;113:9-24. doi: 10.1016/j.semcancer.2025.05.002. Epub 2025 May 8. Semin Cancer Biol. 2025. PMID: 40348000 Review.
-
Isotope tracing in health and disease.Curr Opin Biotechnol. 2022 Aug;76:102739. doi: 10.1016/j.copbio.2022.102739. Epub 2022 Jun 20. Curr Opin Biotechnol. 2022. PMID: 35738210 Free PMC article. Review.
-
13C-Fingerprinting and Metabolic Flux Analysis of Bacterial Metabolisms.Methods Mol Biol. 2019;1927:215-230. doi: 10.1007/978-1-4939-9142-6_15. Methods Mol Biol. 2019. PMID: 30788795
-
Parallel labeling experiments for pathway elucidation and (13)C metabolic flux analysis.Curr Opin Biotechnol. 2015 Dec;36:91-7. doi: 10.1016/j.copbio.2015.08.014. Epub 2015 Aug 28. Curr Opin Biotechnol. 2015. PMID: 26322734 Review.
Cited by
-
Mitochondrial calcium signaling regulates branched-chain amino acid catabolism in fibrolamellar carcinoma.Sci Adv. 2025 May 30;11(22):eadu9512. doi: 10.1126/sciadv.adu9512. Epub 2025 May 28. Sci Adv. 2025. PMID: 40435263 Free PMC article.
-
Mitochondrial Calcium Signaling Regulates Branched-Chain Amino Acid Catabolism in Fibrolamellar Carcinoma.bioRxiv [Preprint]. 2024 Nov 30:2024.05.27.596106. doi: 10.1101/2024.05.27.596106. bioRxiv. 2024. Update in: Sci Adv. 2025 May 30;11(22):eadu9512. doi: 10.1126/sciadv.adu9512. PMID: 38853984 Free PMC article. Updated. Preprint.
-
Polychlorinated Biphenyl Exposure Alters tRNA Transcriptome in High-Fat Diet-Fed Mouse Liver.Noncoding RNA. 2025 May 22;11(3):41. doi: 10.3390/ncrna11030041. Noncoding RNA. 2025. PMID: 40559619 Free PMC article.
References
-
- Felig, P. Amino acid metabolism in man. Annu. Rev. Biochem.44, 933–955 (1975). - PubMed

