TRAP1 modulates mitochondrial biogenesis via PGC-1α/TFAM signalling pathway in colorectal cancer cells
- PMID: 39210159
- PMCID: PMC11416412
- DOI: 10.1007/s00109-024-02479-9
TRAP1 modulates mitochondrial biogenesis via PGC-1α/TFAM signalling pathway in colorectal cancer cells
Abstract
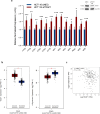
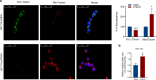
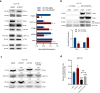
Metabolic rewiring promotes cancer cell adaptation to a hostile microenvironment, representing a hallmark of cancer. This process involves mitochondrial function and is mechanistically linked to the balance between mitochondrial biogenesis (MB) and mitophagy. The molecular chaperone TRAP1 is overexpressed in 60-70% of human colorectal cancers (CRC) and its over-expression correlates with poor clinical outcome, being associated with many cancer cell functions (i.e. adaptation to stress, protection from apoptosis and drug resistance, protein synthesis quality control, metabolic rewiring from glycolysis to mitochondrial respiration and vice versa). Here, the potential new role of TRAP1 in regulating mitochondrial dynamics was investigated in CRC cell lines and human CRCs. Our results revealed an inverse correlation between TRAP1 and mitochondrial-encoded respiratory chain proteins both at transcriptional and translational levels. Furthermore, TRAP1 silencing is associated with increased mitochondrial mass and mitochondrial DNA copy number (mtDNA-CN) as well as enhanced MB through PGC-1α/TFAM signalling pathway, promoting the formation of new functioning mitochondria and, likely, underlying the metabolic shift towards oxidative phosphorylation. These results suggest an involvement of TRAP1 in regulating MB process in human CRC cells. KEY MESSAGES: TRAP1 inversely correlates with protein-coding mitochondrial gene expression in CRC cells and tumours. TRAP1 silencing correlates with increased mitochondrial mass and mtDNA copy number in CRC cells. TRAP1 silencing favours mitochondrial biogenesis in CRC cells.
Keywords: Colorectal cancer; Metabolism; Mitochondrial biogenesis; Peroxisome proliferation-activated receptor gamma coactivator α1-alpha; TNF receptor-associated protein 1; Transcription factor A mitochondrial.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

