Common molecular and pathophysiological underpinnings of delirium and Alzheimer's disease: molecular signatures and therapeutic indications
- PMID: 39210294
- PMCID: PMC11363673
- DOI: 10.1186/s12877-024-05289-3
Common molecular and pathophysiological underpinnings of delirium and Alzheimer's disease: molecular signatures and therapeutic indications
Abstract
Background: Delirium and Alzheimer's disease (AD) are common causes of cognitive dysfunction among older adults. These neurodegenerative diseases share a common and complex relationship, and can occur individually or concurrently, increasing the chance of permanent mental dysfunction. However, the common molecular pathophysiology, key proteomic biomarkers, and functional pathways are largely unknown, whereby delirium is superimposed on AD and dementia.
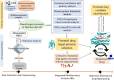
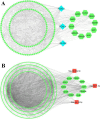
Methods: We employed an integrated bioinformatics and system biology analysis approach to decipher such common key proteomic signatures, pathophysiological links between delirium and AD by analyzing the gene expression data of AD-affected human brain samples and comparing them with delirium-associated proteins. The present study identified the common drug target hub-proteins examining the protein-protein interaction (PPI) and gene regulatory network analysis. The functional enrichment and pathway analysis was conducted to reveal the common pathophysiological relationship. Finally, the molecular docking and dynamic simulation was used to computationally identify and validate the potential drug target and repurposable drugs for delirium and AD.
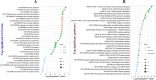
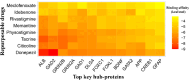
Results: We detected 99 shared differentially expressed genes (sDEGs) associated with AD and delirium. The sDEGs-set enrichment analysis detected the transmission across chemical synapses, neurodegeneration pathways, neuroinflammation and glutamatergic signaling pathway, oxidative stress, and BDNF signaling pathway as the most significant signaling pathways shared by delirium and AD. The disease-sDEGs interaction analysis highlighted the other disease risk factors with delirium and AD development and progression. Among the sDEGs of delirium and AD, the top 10 hub-proteins including ALB, APP, BDNF, CREB1, DLG4, GAD1, GAD2, GFAP, GRIN2B and GRIN2A were found by the PPI network analysis. Based on the maximum molecular docking binding affinities and molecular dynamic simulation (100 ns) results, the ALB and GAD2 were found as prominent drug target proteins when tacrine and donepezil were identified as potential drug candidates for delirium and AD.
Conclusion: The study outlined the common key biomolecules and biological pathways shared by delirium and AD. The computationally reported potential drug molecules need a deeper investigation including clinical trials to validate their effectiveness. The outcomes from this study will help to understand the typical pathophysiological relationship between delirium and AD and flag future therapeutic development research for delirium.
Keywords: Alzheimer’s disease; Common signaling pathways; Delirium; Drug repurposing; Essential drug targets; Molecular docking simulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

