Construction of a prognostic model for disulfidptosis-related long noncoding RNAs in R0 resected hepatocellular carcinoma and analysis of their impact on malignant behavior
- PMID: 39210306
- PMCID: PMC11363604
- DOI: 10.1186/s12885-024-12816-3
Construction of a prognostic model for disulfidptosis-related long noncoding RNAs in R0 resected hepatocellular carcinoma and analysis of their impact on malignant behavior
Abstract
Background: Disulfidptosis is an emerging form of cellular death resulting from the binding of intracellular disulfide bonds to actin cytoskeleton proteins. This study aimed to investigate the expression and prognostic significance of hub disulfidptosis-related lncRNAs (DRLRs) in R0 resected hepatocellular carcinoma (HCC) as well as their impact on the malignant behaviour of HCC cells.
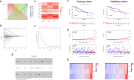
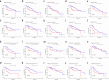
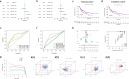
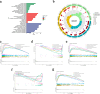
Methods: A robust signature for R0 resected HCC was constructed using least absolute shrinkage and selection operator (LASSO) and multivariate Cox regression and was validated in an independent internal validation cohort to predict the prognosis of R0 HCC patients. Comprehensive bioinformatics analysis was performed on the hub DRLRs (KDM4A-AS1, MKLN1-AS, and TMCC1-AS1), followed by experimental validation using quantitative real-time polymerase chain reaction (qRT‒PCR) and cellular functional assays.
Results: The signature served as an independent prognostic factor applicable to R0 HCC patients across different age groups, tumour stages, and pathological characteristics. Gene Ontology (GO) and gene set enrichment analysis (GSEA) revealed hub pathways associated with this signature. The high-risk group presented an increased abundance of M0 macrophages and activated memory CD4 T cells as well as elevated macrophage and major histocompatibility complex (MHC) class I expression. High-risk R0 HCC patients also presented increased tumour immune dysfunction and exclusion scores (TIDEs), mutation frequencies, and tumour mutational burdens (TMBs). Drug sensitivity analysis revealed that high-risk patients were more responsive to drugs, including GDC0810 and osimertinib. High expression levels of the three hub DRLRs were detected in R0 HCC tissues and HCC cell lines. Functional assays revealed that the three hub DRLRs enhanced HCC cell proliferation, migration, and invasion.
Conclusions: A signature was constructed on the basis of three DRLRs, providing novel insights for personalized precision therapy in R0 HCC patients.
Keywords: Bioinformatics; Disulfidptosis; Hepatocellular carcinoma; KDM4A-AS1; Long noncoding RNA; MKLN1-AS; Malignant biological behaviours; R0 resection; TMCC1-AS1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Based on disulfidptosis, unveiling the prognostic and immunological signatures of Asian hepatocellular carcinoma and identifying the potential therapeutic target ZNF337-AS1.Discov Oncol. 2025 Apr 17;16(1):544. doi: 10.1007/s12672-025-02325-5. Discov Oncol. 2025. PMID: 40244531 Free PMC article.
-
Constructed Risk Prognosis Model Associated with Disulfidptosis lncRNAs in HCC.Int J Mol Sci. 2023 Dec 18;24(24):17626. doi: 10.3390/ijms242417626. Int J Mol Sci. 2023. PMID: 38139458 Free PMC article.
-
A disulfidptosis-related lncRNA signature for analyzing tumor microenvironment and clinical prognosis in hepatocellular carcinoma.Front Immunol. 2024 Oct 7;15:1412277. doi: 10.3389/fimmu.2024.1412277. eCollection 2024. Front Immunol. 2024. PMID: 39434887 Free PMC article.
-
Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma.World J Gastroenterol. 2019 Jan 14;25(2):220-232. doi: 10.3748/wjg.v25.i2.220. World J Gastroenterol. 2019. PMID: 30670911 Free PMC article.
-
Therapeutic Potential of lncRNAs in Regulating Disulfidptosis for Cancer Treatment.Pathol Res Pract. 2024 Nov;263:155657. doi: 10.1016/j.prp.2024.155657. Epub 2024 Oct 16. Pathol Res Pract. 2024. PMID: 39437641 Review.
Cited by
-
Elucidating the role of AC026412.3 in hepatocellular carcinoma: a prognostic disulfidptosis-related LncRNAs model perspective.BMC Gastroenterol. 2025 Aug 12;25(1):579. doi: 10.1186/s12876-025-04174-6. BMC Gastroenterol. 2025. PMID: 40790550 Free PMC article.
-
Long non-coding RNA-based single and combination independent prognostic biomarkers for hepatocellular carcinoma.Discov Oncol. 2025 Sep 2;16(1):1674. doi: 10.1007/s12672-025-03309-1. Discov Oncol. 2025. PMID: 40892177 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

