Integration and validation of host transcript signatures, including a novel 3-transcript tuberculosis signature, to enable one-step multiclass diagnosis of childhood febrile disease
- PMID: 39210372
- PMCID: PMC11360490
- DOI: 10.1186/s12967-024-05241-4
Integration and validation of host transcript signatures, including a novel 3-transcript tuberculosis signature, to enable one-step multiclass diagnosis of childhood febrile disease
Abstract
Background: Whole blood host transcript signatures show great potential for diagnosis of infectious and inflammatory illness, with most published signatures performing binary classification tasks. Barriers to clinical implementation include validation studies, and development of strategies that enable simultaneous, multiclass diagnosis of febrile illness based on gene expression.
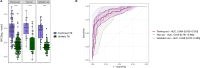
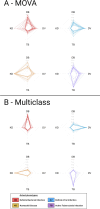
Methods: We validated five distinct diagnostic signatures for paediatric infectious diseases in parallel using a single NanoString nCounter® experiment. We included a novel 3-transcript signature for childhood tuberculosis, and four published signatures which differentiate bacterial infection, viral infection, or Kawasaki disease from other febrile illnesses. Signature performance was assessed using receiver operating characteristic curve statistics. We also explored conceptual frameworks for multiclass diagnostic signatures, including additional transcripts found to be significantly differentially expressed in previous studies. Relaxed, regularised logistic regression models were used to derive two novel multiclass signatures: a mixed One-vs-All model (MOVA), running multiple binomial models in parallel, and a full-multiclass model. In-sample performance of these models was compared using radar-plots and confusion matrix statistics.
Results: Samples from 91 children were included in the study: 23 bacterial infections (DB), 20 viral infections (DV), 14 Kawasaki disease (KD), 18 tuberculosis disease (TB), and 16 healthy controls. The five signatures tested demonstrated cross-platform performance similar to their primary discovery-validation cohorts. The signatures could differentiate: KD from other diseases with area under ROC curve (AUC) of 0.897 [95% confidence interval: 0.822-0.972]; DB from DV with AUC of 0.825 [0.691-0.959] (signature-1) and 0.867 [0.753-0.982] (signature-2); TB from other diseases with AUC of 0.882 [0.787-0.977] (novel signature); TB from healthy children with AUC of 0.910 [0.808-1.000]. Application of signatures outside of their designed context reduced performance. In-sample error rates for the multiclass models were 13.3% for the MOVA model and 0.0% for the full-multiclass model. The MOVA model misclassified DB cases most frequently (18.7%) and TB cases least (2.7%).
Conclusions: Our study demonstrates the feasibility of NanoString technology for cross-platform validation of multiple transcriptomic signatures in parallel. This external cohort validated performance of all five signatures, including a novel sparse TB signature. Two exploratory multi-class models showed high potential accuracy across four distinct diagnostic groups.
Keywords: Bacterial infection; Diagnostics; Gene expression; Kawasaki disease; Multiclass diagnostics; Tuberculosis; Viral infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following filed patents and patent applications, relating to specific transcripts described in this manuscript: 2304229.4/GB/PRV, 23-03-2023; EP4200860A1/WO, 24-08-2021; CA3135429A1, 04-08-2018; ES2941905T3, 12-07-2017.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- MRF-160-0008-ELP-KAFO-C0801/MRF/MRF/United Kingdom
- NIHR/SRF-2009-02-07/National Institute for Health and Care Research
- 10 PXIB 918 184 PR/Promotion of Research Project, Regional Galician funds
- Sante/2006/105-061/EU Action for diseases of poverty program
- WMNP_P69099/NIHR Imperial Biomedical Research Centre
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

