Evaluating the generalizability of graph neural networks for predicting collision cross section
- PMID: 39210378
- PMCID: PMC11363525
- DOI: 10.1186/s13321-024-00899-w
Evaluating the generalizability of graph neural networks for predicting collision cross section
Abstract
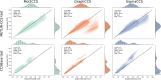
Ion Mobility coupled with Mass Spectrometry (IM-MS) is a promising analytical technique that enhances molecular characterization by measuring collision cross-section (CCS) values, which are indicative of the molecular size and shape. However, the effective application of CCS values in structural analysis is still constrained by the limited availability of experimental data, necessitating the development of accurate machine learning (ML) models for in silico predictions. In this study, we evaluated state-of-the-art Graph Neural Networks (GNNs), trained to predict CCS values using the largest publicly available dataset to date. Although our results confirm the high accuracy of these models within chemical spaces similar to their training environments, their performance significantly declines when applied to structurally novel regions. This discrepancy raises concerns about the reliability of in silico CCS predictions and underscores the need for releasing further publicly available CCS datasets. To mitigate this, we introduce Mol2CCS which demonstrates how generalization can be partially improved by extending models to account for additional features such as molecular fingerprints, descriptors, and the molecule types. Lastly, we also show how confidence models can support by enhancing the reliability of the CCS estimates.Scientific contributionWe have benchmarked state-of-the-art graph neural networks for predicting collision cross section. Our work highlights the accuracy of these models when trained and predicted in similar chemical spaces, but also how their accuracy drops when evaluated in structurally novel regions. Lastly, we conclude by presenting potential approaches to mitigate this issue.
© 2024. The Author(s).
Conflict of interest statement
All authors were employees of Enveda Biosciences Inc. during the course of this work and have real or potential ownership interest in the company.
Figures



References
LinkOut - more resources
Full Text Sources

