Identification of a specific APOE transcript and functional elements associated with Alzheimer's disease
- PMID: 39210471
- PMCID: PMC11361112
- DOI: 10.1186/s13024-024-00751-7
Identification of a specific APOE transcript and functional elements associated with Alzheimer's disease
Abstract
Background: The APOE gene is the strongest genetic risk factor for late-onset Alzheimer's Disease (LOAD). However, the gene regulatory mechanisms at this locus remain incompletely characterized.
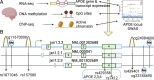
Methods: To identify novel AD-linked functional elements within the APOE locus, we integrated SNP variants with multi-omics data from human postmortem brains including 2,179 RNA-seq samples from 3 brain regions and two ancestries (European and African), 667 DNA methylation samples, and ChIP-seq samples. Additionally, we plotted the expression trajectory of APOE transcripts in human brains during development.
Results: We identified an AD-linked APOE transcript (jxn1.2.2) particularly observed in the dorsolateral prefrontal cortex (DLPFC). The APOE jxn1.2.2 transcript is associated with brain neuropathological features, cognitive impairment, and the presence of the APOE4 allele in DLPFC. We prioritized two independent functional SNPs (rs157580 and rs439401) significantly associated with jxn1.2.2 transcript abundance and DNA methylation levels. These SNPs are located within active chromatin regions and affect brain-related transcription factor-binding affinities. The two SNPs shared effects on the jxn1.2.2 transcript between European and African ethnic groups.
Conclusion: The novel APOE functional elements provide potential therapeutic targets with mechanistic insight into the disease etiology.
Keywords: APOE; Alzheimer’s disease; Postmortem brain; SNP; Transcript.
© 2024. The Author(s).
Conflict of interest statement
All authors declare they have no competing interests.
Figures




Update of
-
Identification of a specific APOE transcript and functional elements associated with Alzheimer's disease.medRxiv [Preprint]. 2023 Oct 31:2023.10.30.23297431. doi: 10.1101/2023.10.30.23297431. medRxiv. 2023. Update in: Mol Neurodegener. 2024 Aug 29;19(1):63. doi: 10.1186/s13024-024-00751-7. PMID: 37961425 Free PMC article. Updated. Preprint.
References
-
- Alzheimer’s_Association. Alzheimer’s disease facts and figures. Alzheimers Dement. 2018;2018:367–429.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

