Immunoproteasome Activation Expands the MHC Class I Immunopeptidome, Unmasks Neoantigens, and Enhances T-cell Anti-Myeloma Activity
- PMID: 39210605
- PMCID: PMC11612626
- DOI: 10.1158/1535-7163.MCT-23-0931
Immunoproteasome Activation Expands the MHC Class I Immunopeptidome, Unmasks Neoantigens, and Enhances T-cell Anti-Myeloma Activity
Abstract
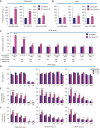
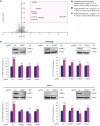
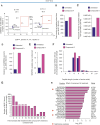
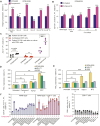
Proteasomes generate antigenic peptides that are presented on the tumor surface to cytotoxic T-lymphocytes. Immunoproteasomes are highly specialized proteasome variants that are expressed at higher levels in antigen-presenting cells and contain replacements of the three constitutive proteasome catalytic subunits to generate peptides with a hydrophobic C-terminus that fit within the groove of MHC class I (MHC-I) molecules. A hallmark of cancer is the ability to evade immunosurveillance by disrupting the antigen presentation machinery and downregulating MHC-I antigen presentation. High-throughput screening was performed to identify compound A, a novel molecule that selectively increased immunoproteasome activity and expanded the number and diversity of MHC-I-bound peptides presented on multiple myeloma cells. Compound A increased the presentation of individual MHC-I-bound peptides by >100-fold and unmasked tumor-specific neoantigens on myeloma cells. Global proteomic integral stability assays determined that compound A binds to the proteasome structural subunit PSMA1 and promotes association of the proteasome activator PA28α/β (PSME1/PSME2) with immunoproteasomes. CRISPR/Cas9 silencing of PSMA1, PSME1, or PSME2 as well as treatment with immunoproteasome-specific suicide inhibitors abolished the effects of compound A on antigen presentation. Treatment of multiple myeloma cell lines and patient bone marrow-derived CD138+ cells with compound A increased the anti-myeloma activity of allogenic and autologous T cells. Compound A was well-tolerated in vivo and co-treatment with allogeneic T cells reduced the growth of myeloma xenotransplants in NOD/SCID gamma mice. Taken together, our results demonstrate the paradigm shifting impact of immunoproteasome activators to diversify the antigenic landscape, expand the immunopeptidome, potentiate T-cell-directed therapy, and reveal actionable neoantigens for personalized T-cell immunotherapy.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
E. Malek reports being on the advisory board for Bristol Myers Squibb, Janssen, and Amgen; research funding from MedPacto Inc.; and being a speaker for Bristol Myers Squibb and Janssen. D.J. Adams reports personal fees and other support from Convelo Therapeutics outside the submitted work. J.J. Driscoll reports a patent for UH Ventures pending. No disclosures were reported by the other authors.
Figures



References
-
- Yang K, Halima A, Chan TA. Antigen presentation in cancer—mechanisms and clinical implications for immunotherapy. Nat Rev Clin Oncol 2023;20:604–23. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

