Evaluating Imide-Based Mass Spectrometry-Cleavable Cross-Linkers for Structural Proteomics Studies
- PMID: 39211594
- PMCID: PMC11350583
- DOI: 10.1021/jacsau.4c00282
Evaluating Imide-Based Mass Spectrometry-Cleavable Cross-Linkers for Structural Proteomics Studies
Abstract
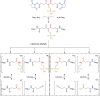
Disuccinimidyl dibutyric urea (DSBU) is a mass spectrometry (MS)-cleavable cross-linker that has multiple applications in structural biology, ranging from isolated protein complexes to comprehensive system-wide interactomics. DSBU facilitates a rapid and reliable identification of cross-links through the dissociation of its urea group in the gas phase. In this study, we further advance the structural capabilities of DSBU by remodeling the urea group into an imide, thus introducing a novel class of cross-linkers. This modification preserves the MS cleavability of the amide bond, granted by the two acyl groups of the imide function. The central nitrogen atom enables the introduction of affinity purification tags. Here, we introduce disuccinimidyl disuccinic imide (DSSI) as a prototype of this class of cross-linkers. It features a phosphonate handle for immobilized metal ion affinity chromatography enrichment. We detail DSSI synthesis and describe its behavior in solution and in the gas phase while cross-linking isolated proteins and human cell lysates. DSSI and DSBU cross-links are compared at the same enrichment depth to bridge these two cross-linker classes. We validate DSSI cross-links by mapping them in high-resolution structures of large protein assemblies. The cross-links observed yield insights into the morphology of intrinsically disordered proteins and their complexes. The DSSI linker might spearhead a novel class of MS-cleavable and enrichable cross-linkers.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
LinkOut - more resources
Full Text Sources
